Introduction
MicroRNAs (miRNAs), non-coding RNAs of ~22 nt in
length, act as post-transcriptional regulators. miRNAs control mRNA
stability and the efficiency of translation by base pairing with
complementary sites in the 3′-untranslated region of mRNA (1). The most recent release of miRBase
(release 20; http://microrna.sanger.ac.uk/) annotates 2,578 miRNA
loci in the human genome (2). A class
of these miRNAs has been associated with human cancer (3–6) and are
referred to as ‘oncomirs’ (7).
Oncomirs are divided into two groups: i) miRNAs that are
upregulated or amplified in cancer and are likely to act as
oncogenes; and ii) miRNAs that are deleted or downregulated in
cancer and are likely to act as tumor suppressors. Aberrant
expression of miRNAs is associated with proliferation (8), invasion (9), apoptosis (10) and signaling pathways (11) in the progression of breast cancer.
Considering the association of miRNAs with cancer development and
progression, it has been suggested that these small regulatory RNAs
serve as potential targets of anticancer therapeutic strategies
(12).
Breast cancer is one of the most common types of
cancer and has the highest cancer-specific mortality rate in women
worldwide (13). It is therefore
essential to better understand the underlying molecular mechanisms
and to develop novel approaches for the prevention, treatment, and
management of breast cancer. Currently, treatment strategy for
breast cancer include locoregional treatment with surgery and
radiation, plus systemic treatment with chemotherapy, endocrine,
and biologic therapies (13).
Considering the association of miRNAs with cancer development and
progression, it has been suggested that these small regulatory RNAs
serve as potential targets of anticancer therapeutic strategies
(14).
Cantharidin (CTD) is one of various natural products
used in traditional Chinese medicine for the treatment of cancer.
CTD can effectively inhibit the proliferation, break the DNA
strands of human CCRF-CEM leukemia cells (14), reverse multidrug resistance of
hepatoma HepG2/ADM cells (15) and
induce apoptosis of human multiple myeloma cells (RPMI-8226, U266,
and IM9) (16). However, the exact
anti-cancer mechanism of CTD in human cancer cells remains poorly
understood. Combined methods of pharmaceutical biology and
molecular biology may facilitate the elucidation of the modes of
action of these natural products.
In the present study, a microarray chip was used for
miRNA expression profiling to test our hypothesis that CTD alters
miRNA expression profiles in breast cancer cells. The results
indicated that CTD alters specific miRNA expression in human breast
cancer cells. Furthermore, gene expression that is targeted by
these specific miRNAs downregulated by CTD was also modulated by
CTD. It therefore appears that miRNA-related changes are an
important effect of CTD.
Materials and methods
Cell culture
MCF-7, MDA-MB-231 and HBL-100 human breast cancer
cells were obtained from the American Type Culture Collection
(Manassas, VA, USA) and cultured in RPMI-1640 medium (Gibco, San
Francisco, CA, USA) supplemented with 10% heat-inactivated (56°C,
30 min) fetal calf serum (A-4061; PAA Laboratories GmbH, Pasching,
Austria), 0.01 mg/ml insulin (Sigma-Aldrich, St. Louis, MO, USA), 2
mM L-glutamine (Gibco), 100 U/ml penicillin and 100 µg/ml
streptomycin. The cell culture was maintained at 37°C in a 5%
CO2 humidified atmosphere.
CTD treatment and cell proliferation
assay
MCF-7 cells were seeded in 96-well culture plates
(104 cells/well). After overnight incubation, various
concentrations of CTD (Nanjing Pharmaceutical Factory Co., Ltd.,
Nanjing China) dissolved in dimethyl sulfoxide (DMSO;
Sigma-Aldrich) were added to the plates (0.0, 0.8, 1.6, 3.2, 6.4
and 12.8 µg/ml). DMSO was adjusted to the same final concentration
of 0.01%. Following incubation, cell growth was measured by the
addition of 20 µl 5 g/l MTT (Gen-View Scientific, Inc., Calimesa,
CA, USA) at 37°C for 4 h. DMSO (150 µl) was then added to dissolve
the formazan crystals. Absorbance was measured at 490 nm using an
enzyme-linked immunosorbent assay plate reader (BioTek, Winooski,
VT, USA) to obtain optical density (OD) values. The percentage of
inhibition was calculated as: Inhibition ratio (IR) = (1 -
ODsample / ODcontrol) × 100%. The
IC50 (concentration of CTD required to produce 50% cell
inhibition) was determined and used for subsequent analyses.
Experiments were performed in triplicate.
RNA isolation and qualitative
detection
Briefly, MCF-7, MDA-MB-231 and HBL-100 cells were
collected and washed with cold phosphate-buffered saline. Total RNA
was isolated from 5×106 cells using TRIzol® reagent
(Invitrogen Life Technologies, Inc., Carlsbad, CA, USA), according
to the manufacturer's instructions. Absorbance each RNA sample
(MCF-7, MDA-MB-231 and HBL-100 cells) was measured using a NanoDrop
2000 spectrophotometer (ThermoFisher Scientific Inc., Waltham, MA,
USA) at wavelengths of 230, 260 and 280 nm, respectively, to
determine the purification and concentration of the total RNA
samples. The ratio of 28S to 18S in the total RNA sample was
detected by electrophoresis on a 1% agarose gel containing
formaldehyde to evaluate its purification and integrity. RNA
samples conforming to the quality requirements (260/230 nm
intensity ratio >1.0 and 260 /280 nm ratio >1.8; the 28S rRNA
band at 4.5 kb should be twice the intensity of the 18S rRNA band
at 1.9 kb) were used for the microarray assays.
μParaflo™ miRNA microarray assay
A microarray assay was performed using a service
provider (LC Sciences, LLC, Houston, TX, USA). A sample of total
RNA (5 µg) was size-fractionated using a YM-100 Microcon®
centrifugal filter (EMD Millipore, Billerica, MA, USA). The
isolated small RNAs (<300 nt) were 3-extended with a polyA
polymerase (Invitrogen Life Technologies). An oligonucleotide tag
was then ligated to the polyA tail. Hybridization was performed
overnight on a µParaflo microfluidic chip using a micro-circulation
pump (Atactic Technologies, Houston, TX, USA). On the microfluidic
chip containing 723 mature human miRNA probes, each detection probe
consisted of a chemically modified nucleotide coding segment
complementary to target miRNA from the miRBase Sequence database
version 10.0 (Wellcome Trust Sanger Institute, Cambridge, UK;
http://microrna.sanger.ac.uk/sequences). The detection
probes were constructed by in situ synthesis using
photogenerated reagent chemistry. Each probe sequence was repeated
five times on the same array chip. The hybridization melting
temperatures were balanced by chemical modifications of the
detection probes. Hybridization was performed overnight using 100
µl 6X SSPE buffer (0.90 M NaCl, 60 mM
Na2HPO4, 6 mM EDTA; pH 6.8) containing 25%
formamide at 34°C. Subsequently, hybridization detection was
performed by fluorescence labeling using tag-specific Cy3 and Cy5
dyes (PerkinElmer, Chalfont, UK). Hybridization images were
obtained using a laser scanner [GenePix® 4000B; Molecular Devices,
LLC, Sunnyvale, CA, USA] and digitized using Array-Pro image
analysis software (Media Cybernetics, Inc., Rockville, MD,
USA).
Microarray data handling and
analysis
The data were analyzed after subtracting the
background and normalizing the signals using a LOWESS filter
(locally-weighted regression). For two-color experiments, the ratio
of the two sets of detected signals (log2 transformed,
balanced) and P-values of the t-test were calculated.
P<0.01 were considered to indicate a statistically significant
difference and a fold-change of ≥2 was regarded as statistically
significant.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR) for miRNA analysis
miRNAs, E2F1, MCM7 and GAPDH expression were
measured by RT-qPCR. miRNA analysis was performed using a
Bulge-Loop™ miRNA qRT-PCR primer set (Guangzhou RiboBio Co., Ltd.,
Guangzhou, China), according to the manufacturers instructions. The
U6 small nuclear RNA was used as an internal control for the
miRNAs. The mRNA expression of E2F1, MCM7 was normalized to the
expression of GAPDH. The relative expression levels were performed
with the comparative CT method. Reaction conditions were as
follows: 95°C for 1 min, followed by 40 cycles of 95°C for 15 sec,
60°C for 20 sec, and 72°C for 15 sec. The primers used for E2F1,
MCM7 and GAPDH were as follows: Forward, 5-CCATCCAGGAAAAGGTGTGAA-3
and reverse, 5-AGCGCTTGGTGGTCAGATTC-3′ for E2F1; forward,
5-TCTGGCACGTCTGAGAATGGT-3′ and reverse,
5-ACGGACGGTGGCAAATATCA-3 for MCM7; and forward,
5-AGCCTCAAGATCATCAGCAATG-3 and reverse,
5-CACGATACCAAAGTTGTCATGGAT-3′ for GAPDH. Quantitative
expression data were acquired and analyzed using a LightCycler 1.5
Instrument (Roche Applied Science, Indianapolis, IN, USA).
Transient miRNA transfection
MCF-7 cells at 50% confluency were transfected with
miR-Ribo™ miR-106b inhibitor and negative control inhibitor
(Guangzhou RiboBio Co., Ltd.) to a final concentration of 100
nmol/l with Lipofectamine™ 2000 transfection reagent (Invitrogen
Life Technologies, Inc.), according to the manufacturers
instructions. After 6 h of transfection, the cells were placed in
complete medium and maintained at 37°C in a 5% CO2
atmosphere. Following 48 h of transfection, and RNA was harvested
using TRIzol reagent (BioDev-Tech Co., Beijing, China).
Western blot analysis
Briefly, cell pellets were resuspended in lysis
buffer containing 2 M sodium chloride, 10% NP-40, 10% SDS, 1 M/l
Tris-Cl, 1 g/l phenyl-methylsulfonyl fluoride, 0.1 g/l aprotinin
and 0.01 g/l leupeptin, and incubated at 4°C for 30 min. After
13,400 × g centrifugation for 5 min at 4°C, the protein content of
the supernatant was determined using a protein assay reagent
(Bio-Rad Laboratories, Inc., Hercules, CA, USA). Proteins were
separated on a 10% SDS-PAGE gel and blotted onto polyvinylidene
difluoride membrane. The membrane was blocked with bovine serum
albumin for ~1 h at room temperature. The protein expression was
detected using primary antibody [p21, cat no. 2946 and phosphatase
and tensin homolog (PTEN), cat no. 9559S; 1:1000 dilution; Cell
Signaling Technology, Inc., Beverly, MA, USA] and secondary
antibody (1:800 dilution) conjugated with horseradish peroxidase,
followed by treatment with electrochemiluminescence reagents (GE
Healthcare, Amersham, UK). Images of the western blot films were
acquired using AlphaEase™ (Alpha Innotech Corporation, CA, USA) and
FluorChem™ FC2 software (Medford, NJ, USA).
Statistical analysis
Data were presented as the mean ± standard
deviation. Comparisons between groups were performed by using the
Student's t-test and the statistical analyses were performed
using SPSS software (version 19.0; IBM SPSS, Armonk, NY, USA).
Tests were two-tailed and P<0.01 was considered to indicate a
statistically significant difference.
Results
CTD treatment inhibits the
proliferation of MCF-7 cells
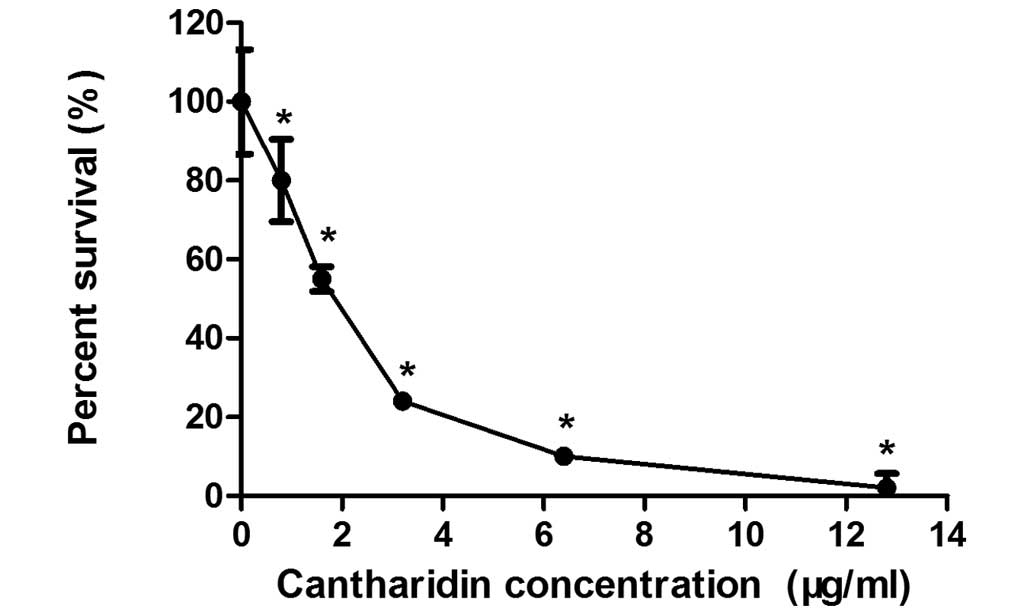
The MTT assay was used to determine the effects of
CTD on MCF-7 human breast cancer cell growth. Fig. 1 shows the effects of 0.8–12.8 µg/ml
CTD on the growth of MCF-7 cells. After 48 h of incubation, CTD
significantly inhibited the proliferation of MCF-7 cells in a
dose-dependent manner (P<0.01). Treatment with CTD
concentrations of 0.8 µg/ml compared with 12.8 µg/ml resulted in a
reduction in cell numbers from 98±4 to 20±7%. The IC50
of CTD was 1.75 µg/ml following treatment for 48 h. Thus, a dose of
1.75 µg/ml CTD was used as the therapeutic drug concentration in
all subsequent experiments.
CTD treatment alters miRNA expression
profiles
To assess whether miRNA expression responds to CTD
treatment in breast cancer, microarray analysis was conducted with
miRNA-enriched total RNAs isolated from MCF-7 cells treated with
1.75 µg/ml CTD for 48 h. RNA samples were processed, labeled and
hybridized to miRNA chips, as described in Materials and methods.
According to the microarray-based screening, changes in miRNA
expression were observed between CTD-treated cells and untreated
control cells. Compared with the control group, Table I indicates that there were 80
differentially expressed miRNAs in the CTD-treated group. Of these,
35 miRNAs were significantly upregulated and 45 miRNAs were
significantly downregulated (P<0.01). A greater number of miRNAs
were downregulated than upregulated following CTD treatment.
 | Table I.Effect of cantharidin on miRNA
expression in MCF-7 cells. |
Table I.
Effect of cantharidin on miRNA
expression in MCF-7 cells.
| miRNA name | Fold change | P-value |
|---|
| Upregulated
(n=35) |
|
hsa-miR-122 | 298.40 | 1.84E-06 |
|
hsa-miR-200c | 50.31 | 2.53E-08 |
|
hsa-miR-936 | 33.44 | 1.74E-05 |
|
hsa-miR-374a | 12.16 | 1.81E-04 |
|
hsa-miR-214 | 8.43 | 4.43E-03 |
|
hsa-miR-149a | 7.90 | 1.05E-06 |
|
hsa-miR-637 | 7.73 | 5.46E-04 |
|
hsa-miR-654-5p | 7.53 | 3.89E-03 |
|
hsa-miR-32a | 7.07 | 6.02E-03 |
|
hsa-miR-198 | 6.68 | 1.88E-03 |
|
hsa-miR-192 | 5.79 | 1.01E-04 |
|
hsa-miR-7 | 5.40 | 2.70E-05 |
|
hsa-miR-194 | 4.94 | 1.88E-03 |
|
hsa-miR-221a | 4.87 | 7.26E-03 |
|
hsa-miR-939 | 4.45 | 7.43E-03 |
|
hsa-miR-96 | 4.03 | 7.57E-03 |
|
hsa-miR-199a-3p | 4.07 | 1.68E-03 |
|
hsa-miR-574-5p | 3.91 | 1.81E-04 |
|
hsa-miR-98 | 3.86 | 1.70E-07 |
|
hsa-miR-483-5p | 3.82 | 1.60E-04 |
|
hsa-miR-139-5p | 3.69 | 4.99E-03 |
|
hsa-miR-22a | 3.50 | 6.17E-05 |
|
hsa-miR-203 | 3.48 | 5.86E-03 |
|
hsa-miR-196a | 3.34 | 6.69E-07 |
|
hsa-miR-148a | 3.17 | 2.16E-03 |
|
hsa-miR-195 | 3.08 | 1.14E-03 |
|
hsa-miR-658 | 3.00 | 3.43E-05 |
|
hsa-miR-663 | 2.92 | 8.74E-05 |
|
hsa-miR-196b | 2.84 | 6.04E-05 |
|
hsa-miR-7-1a | 2.75 | 2.80E-03 |
|
hsa-miR-671-5p | 2.51 | 1.71E-04 |
|
hsa-miR-628-5p | 2.50 | 1.72E-03 |
|
hsa-miR-34c-3p | 2.39 | 1.86E-03 |
|
hsa-miR-638 | 2.30 | 1.10E-05 |
|
hsa-miR-574-3p | 2.26 | 1.58E-03 |
| Downregulated
(n=45) |
|
hsa-miR-801 | 0.50 | 8.94E-05 |
|
hsa-miR-421 | 0.49 | 7.45E-03 |
|
hsa-miR-15a | 0.48 | 4.97E-05 |
|
hsa-miR-193a-5p | 0.45 | 9.49E-07 |
|
hsa-miR-766 | 0.45 | 1.02E-03 |
|
hsa-miR-185 | 0.45 | 2.57E-07 |
|
hsa-miR-138 | 0.43 | 2.80E-03 |
|
hsa-miR-181b | 0.43 | 7.66E-06 |
|
hsa-miR-181a | 0.42 | 1.69E-08 |
|
hsa-miR-30d | 0.42 | 1.12E-06 |
|
hsa-miR-30a | 0.41 | 4.47E-06 |
|
hsa-miR-140-3p | 0.41 | 2.21E-05 |
|
hsa-miR-30aa | 0.40 | 4.92E-06 |
|
hsa-miR-151-3p | 0.40 | 6.72E-07 |
|
hsa-miR-500a | 0.38 | 4.61E-04 |
|
hsa-miR-505a | 0.38 | 3.65E-04 |
|
hsa-miR-532-5p | 0.37 | 6.10E-10 |
|
hsa-miR-25a | 0.36 | 2.93E-03 |
|
hsa-miR-31 | 0.35 | 4.48E-07 |
|
hsa-miR-130b | 0.34 | 1.02E-06 |
|
hsa-miR-362-5p | 0.33 | 1.75E-06 |
|
hsa-miR-503 | 0.33 | 3.24E-06 |
|
hsa-miR-362-3p | 0.33 | 6.24E-05 |
|
hsa-miR-93 | 0.32 | 5.39E-11 |
|
hsa-miR-30c-2a | 0.32 | 2.08E-05 |
|
hsa-miR-423-5p | 0.32 | 5.78E-07 |
|
hsa-miR-193ba | 0.31 | 2.28E-04 |
|
hsa-miR-106b | 0.31 | 9.36E-08 |
|
hsa-miR-744 | 0.30 | 4.02E-06 |
|
hsa-miR-324-5p | 0.29 | 4.05E-07 |
|
hsa-miR-378 | 0.28 | 1.76E-06 |
|
hsa-miR-455-3p | 0.28 | 4.37E-09 |
|
hsa-miR-103 | 0.28 | 1.02E-08 |
|
hsa-miR-18b | 0.27 | 7.63E-05 |
|
hsa-miR-107 | 0.27 | 5.78E-08 |
|
hsa-miR-550a | 0.26 | 2.32E-08 |
|
hsa-miR-877 | 0.25 | 2.93E-04 |
|
hsa-miR-425 | 0.24 | 5.74E-08 |
|
hsa-miR-484 | 0.22 | 4.01E-05 |
|
hsa-miR-34a | 0.21 | 6.82E-08 |
|
hsa-miR-99b | 0.20 | 3.58E-08 |
|
hsa-miR-18a | 0.18 | 4.57E-08 |
|
hsa-miR-197 | 0.16 | 1.29E-08 |
|
hsa-miR-186a | 0.04 | 3.28E-08 |
|
hsa-miR-542-5p | 0.02 | 1.51E-04 |
Validation of microarray results by
RT-qPCR
To validate the microarray data, miR-18a, miR-106b,
miR-93, miR-936 and miR-7 were selected at random and their
expression levels were assayed by RT-qPCR. The results from the
microarray and RT-qPCR were compared. Of the miRNAs selected for
comparison, two miRNAs (miR-936 and miR-7) were significantly
upregulated and three miRNAs (miR-106b, miR-93 and miR-18a) were
significantly downregulated compared with the untreated control
cells based on the results of microarray analysis (P<0.01). The
expression data obtained by RT-qPCR analysis are comparable to the
microarray analysis data, although miR-936 was upregulated to a
lesser degree in the RT-qPCR analysis (Fig. 2).
CTD treatment downregulates the
expression of the miR- 106b-93 host gene MCM7 and its transcription
factor E2F1 in MCF-7 cells
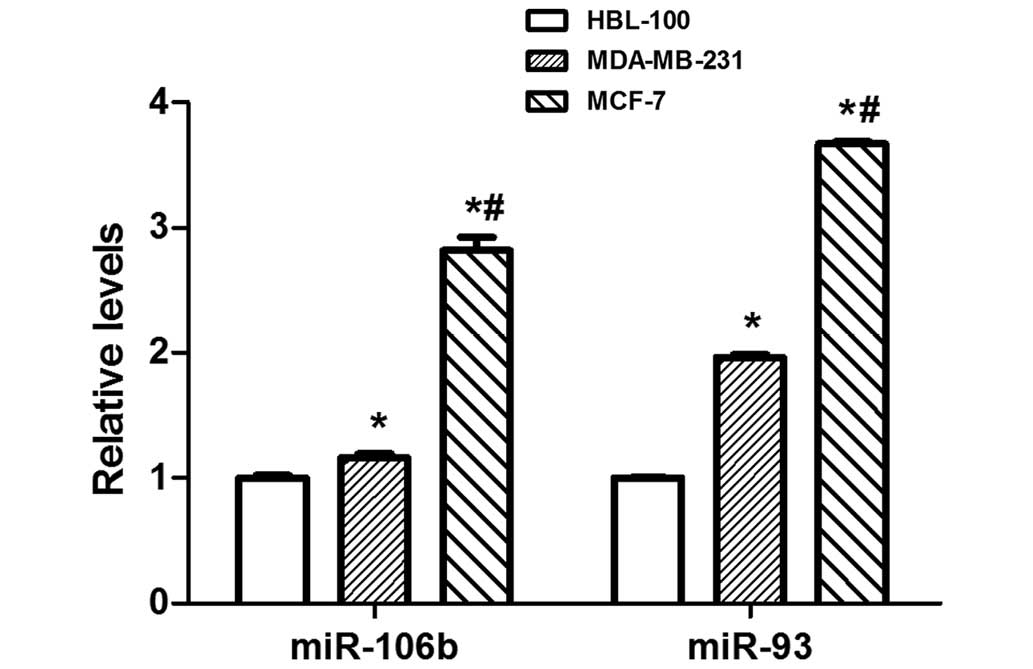
miR-106b-93 are aberrantly overexpressed in numerous
types of human cancer (18–21). Therefore, miR-106b-93 expression was
detected in MCF-7, MDA-MB-231 and HBL-100 cells, and it was
identified that miR-106b-93 expression is significantly elevated in
MCF-7 breast cancer cells compared with MDA-MB-231 and HBL-100
cells (P<0.01; Fig. 3). To
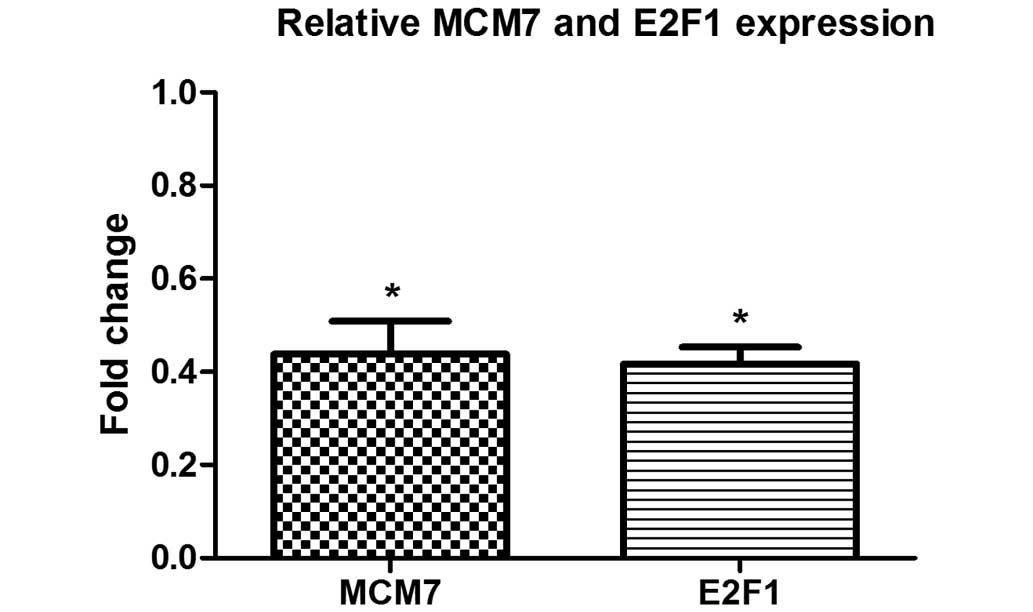
demonstrate the effect of CTD on the expression of miR-106b-93,
host gene MCM7 and its transcription factor E2F1, qPCR was
performed following 48 h of CTD treatment. The results demonstrated
a significant reduction in MCM7 and E2F1 expression following CTD
treatment compared with the untreated control cells (P<0.01;
Fig. 4).
CTD represses miR-106b-93 expression
and induces the protein expression of its targets p21 and PTEN
To demonstrate the effect of CTD on miR-106b-93
target genes p21 and PTEN, western blot analysis was performed
following 48 h CTD treatment. p21 and PTEN have been shown to be
miR-106b and miR-93 post-transcriptional targets (18,20,22). MCF-7
cells exhibited a significant reduction in E2F1, MCM7, miR-106b and
miR-93 expression following CTD treatment (P<0.01; Figs. 2 and 4).
Following confirmation of miR-106b inhibition (Fig. 5A), p21 and PTEN protein expression
were observed to be increased in MCF-7 cells treated with CTD
(Fig. 5B). These results suggested
that CTD inhibits miR-106b and miR-93 expression, and functions in
breast cancer, as indicated by a reduction of its targets, p21 and
PTEN, which are important tumor suppressors in breast cancer.
Discussion
miRNAs are small regulatory RNA molecules that
modulate the expression levels of specific target genes (17). miRNAs function in transcriptional and
post-transcriptional regulation of gene expression; for example,
acting as transcription factors (18). In cancer, miRNAs function as
regulatory molecules, serving as oncogenes or tumor suppressors.
Thus, miRNAs may serve as novel therapeutic targets in cancer
(19).
CTD is one of numerous natural products used in
traditional Chinese medicine for the treatment of cancer. Previous
studies have demonstrated that CTD has potent growth-inhibitory and
proapoptotic effects on cancer. These effects may be mediated
through any of the various mechanisms via which CTD interferes with
cell signaling (14–16). However, the molecular basis of the
effects of CTD remains to be elucidated. In the present study, the
potential modulation of miRNA by CTD was explored. A microarray
chip was used to investigate whether CTD alters miRNA expression
profiles. It was determined that the expressions of 80 miRNAs (fold
change, ≥2; P<0.01) were regulated by CTD exposure in MCF-7
cells. By performing a search of the literature on Pubmed, the
present study identified that numerous breast cancer-related miRNAs
were significantly affected by CTD exposure. For example, previous
studies determined that the expressions of miR-7, miR-195, miR-203
and miR-214 tumor suppressors were downregulated in human breast
cancer (20–23). In the present study, miR-7, miR-195,
miR-203 and miR-214 were significantly upregulated in MCF-7 breast
cancer cells following treatment with CTD. Similarly, the
expression of miR-93, miR-106b and miR-378 were significantly
downregulated by CTD and upregulated in human breast cancer
(24,25). Thus, CTD-induced miRNAs exhibit
anti-tumor activity against breast cancer.
miR-106b-93, which reside in the thirteenth intron
of the MCM7 gene (chromosome 7), is overexpressed in various
types of cancer, including breast (24), hepatocellular (26), gastric (27), and head and neck squamous cell
(28) carcinomas, acting as an
oncogene. miR-106b-93 are considered to be important in cancer
progression by targeting tumor suppressor genes CDKN1A (p21)
and PTEN (27,29). Furthermore, miR-106b-93 are activated
by E2F1 in parallel with its host gene, MCM7 (30). In turn, miR-106b and miR-93 regulate
E2F1 expression, establishing a miRNA negative feedback loop
(30). E2F1 functions as an oncogenic
transcription factor to promote breast cancer cell proliferation
and its expression has been observed to be significantly increased
in breast cancer (31). In the
present study, it was identified that MCF-7 cells exhibit a
significant reduction in E2F1, MCM7, miR-106b and miR-93 expression
following CTD treatment (Figs. 2 and
4). Furthermore, the protein
expression of miR-106b-93 target genes p21 and PTEN
were markedly increased in MCF-7 cells following CTD treatment
(Fig. 5). These results indicate that
CTD decreases transcription factor E2F1 levels, represses the
transactivation of the MCM7 gene, miR-106b-93 expression, and
enhances the expression of the miR-106b-93 targets p21 and
PTEN.
In conclusion, increasing evidence indicates the
role of miRNAs as oncogenes and/or tumor suppressor genes within
the gene regulatory networks. The contribution of miRNAs to the
development of cancer has significant implications in the future of
personalized medicine and cancer treatment. For example, miRNAs may
serve as diagnostic and prognostic markers, therapeutic targets or
tools in cancer diagnosis and treatment. The present study
identified a significant upregulation of tumor-suppressing miRNAs
and a significant downregulation of oncogenic miRNAs following CTD
treatment of MCF-7 cells. In particular, CTD may affect the
E2F1/MCM7-miR-106b-93/p21-PTEN signaling pathway. This result
suggests an important and novel mechanism by which CTD mediates its
potent effects on cell growth and proliferation. However, the
results obtained in this study remain to be verified in future
investigations.
Acknowledgements
The present study was supported by the National
Natural Science Foundation of China (no. 81373518), as well as
grants from the Doctoral Fund of Ministry of Education of China
(grant no. 20093107120010) and Xinglin Scholars Projects of
Shanghai University of Traditional Chinese Medicine (grant nos.
R13010109 and A1-20141009).
References
|
1
|
Filipowicz W, Bhattacharyya SN and
Sonenberg N: Mechanisms of post-transcriptional regulation by
microRNAs: Are the answers in sight? Nat Rev Genet. 9:102–114.
2008. View
Article : Google Scholar : PubMed/NCBI
|
|
2
|
Griffiths-Jones S, Saini HK, van Dongen S
and Enright AJ: miRBase: Tools for microRNA genomics. Nucleic Acids
Res. 36:D154–D158. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Johnson SM, Grosshans H, Shingara J, Byrom
M, Jarvis R, Cheng A, Labourier E, Reinert KL, Brown D and Slack
FJ: RAS is regulated by the let-7 microRNA family. Cell.
120:635–647. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bonci D, Coppola V, Musumeci M, Addario A,
Giuffrida R, Memeo L, D'Urso L, Pagliuca A, Biffoni M, Labbaye C,
et al: The miR-15a-miR-16-1 cluster controls prostate cancer by
targeting multiple oncogenic activities. Nat Med. 14:1271–1277.
2008. View
Article : Google Scholar : PubMed/NCBI
|
|
5
|
Si ML, Zhu S, Wu H, Lu Z, Wu F and Mo YY:
MiR-21-mediated tumor growth. Oncogene. 26:2799–2803. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lee DY, Deng Z, Wang CH and Yang BB:
MicroRNA-378 promotes cell survival, tumor growth and angiogenesis
by targeting SuFu and Fus-1 expression. Proc Natl Acad Sci (USA).
104:20350–20355. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Esquela-Kerscher A and Slack FJ: Oncomirs:
microRNAs with a role in cancer. Nat Rev Cancer. 6:259–269. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hossain A, Kuo MT and Saunders GF:
Mir-17-5p regulates breast cancer cell proliferation by inhibiting
translation of AIB1 mRNA. Mol Cell Biol. 26:8191–8201. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ma L, Teruya-Feldstein J and Weinberg RA:
Tumour invasion and metastasis initiated by microRNA-10b in breast
cancer. Nature. 449:682–688. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Frankel LB, Christoffersen NR, Jacobsen A,
Lindow M, Krogh A and Lund AH: Programmed cell death 4 (PDCD4) is
an important functional target of the microRNA miR-21 in breast
cancer cells. J Biol Chem. 283:1026–1033. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wang V and Wu W: MicroRNA: A new player in
breast cancer development (Review). Cancer Mol. 3:133–138.
2007.
|
|
12
|
Liu G, Wong-Staal F and Li QX: Development
of new RNAi therapeutics. Histol Histopathol. 22:211–217.
2007.PubMed/NCBI
|
|
13
|
Miller BA, Chu KC, Hankey BF and Ries LA:
Cancer incidence and mortality patterns among specific Asian and
pacific islander populations in the U.S. Cancer Causes Control.
19:227–256. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Efferth T, Rauh R, Kahl S, Tomicic M,
Böchzelt H, Tome ME, Briehl MM, Bauer R and Kaina B: Molecular
modes of action of cantharidin in tumor cells. Biochem Pharmacol.
69:811–818. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zheng LH, Bao YL, Wu Y, Yu CL, Meng X and
Li YX: Cantharidin reverses multidrug resistance of human hepatoma
HepG2/ADM cells via down-regulation of P-glycoprotein expression.
Cancer Lett. 272:102–109. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Sagawa M, Nakazato T, Uchida H, Ikeda Y
and Kizaki M: Cantharidin induces apoptosis of human multiple
myeloma cells via inhibition of the JAK/STAT pathway. Cancer Sci.
99:1820–1826. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Shivdasani RA: MicroRNAs: Regulators of
gene expression and cell differentiation. Blood. 108:3646–3653.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ross JS, Carlson JA and Brock G: miRNA:
The new gene silencer. Am J Clin Pathol. 128:830–836. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Chen CZ: MicroRNAs as oncogenes and tumor
suppressors. N Engl J Med. 353:1768–1771. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Reddy SD, Ohshiro K, Rayala SK and Kumar
R: MicroRNA-7, a homeobox D10 target, inhibits p21-activated kinase
1 and regulates its functions. Cancer Res. 68:8195–8200. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Li D, Zhao Y, Liu C, Chen X, Qi Y, Jiang
Y, Zou C, Zhang X, Liu S, Wang X, et al: Analysis of MiR-195 and
MiR-497 expression, regulation and role in breast cancer. Clin
Cancer Res. 17:1722–1730. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wang C, Zheng X, Shen C and Shi Y:
MicroRNA-203 suppresses cell proliferation and migration by
targeting BIRC5 and LASP1 in human triple-negative breast cancer
cells. J Exp Clin Cancer Res. 31:582012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Derfoul A, Juan AH, Difilippantonio MJ,
Palanisamy N, Ried T and Sartorelli V: Decreased microRNA-214
levels in breast cancer cells coincides with increased cell
proliferation, invasion and accumulation of the Polycomb Ezh2
methyltransferase. Carcinogenesis. 32:1607–1614. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ivanovska I, Ball AS, Diaz RL, Magnus JF,
Kibukawa M, Schelter JM, Kobayashi SV, Lim L, Burchard J, Jackson
AL, et al: MicroRNAs in the miR-106b family regulate p21/CDKN1A and
promote cell cycle progression. Mol Cell Biol. 28:2167–2174. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Feng M, Li Z, Aau M, Wong CH, Yang X and
Yu Q: Myc/miR-378/TOB2/cyclin D1 functional module regulates
oncogenic transformation. Oncogene. 30:2242–2251. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Li Y, Tan W, Neo TW, Aung MO, Wasser S,
Lim SG and Tan TM: Role of the miR-106b-25 microRNA cluster in
hepatocellular carcinoma. Cancer Sci. 100:1234–1242. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kan T, Sato F, Ito T, Matsumura N, David
S, Cheng Y, Agarwal R, Paun BC, Jin Z, Olaru AV, et al: The
miR-106b-25 polycistron, activated by genomic amplification,
functions as an oncogene by suppressing p21 and Bim.
Gastroenterology. 136:1689–1700. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Hui AB, Lenarduzzi M, Krushel T, Waldron
L, Pintilie M, Shi W, Perez-Ordonez B, Jurisica I, O'Sullivan B,
Waldron J, et al: Comprehensive MicroRNA profiling for head and
neck squamous cell carcinomas. Clin Cancer Res. 16:1129–1139. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Poliseno L, Salmena L, Riccardi L, Fornari
A, Song MS, Hobbs RM, Sportoletti P, Varmeh S, Egia A, Fedele G, et
al: Identification of the miR-106b~25 microRNA cluster as a
proto-oncogenic PTEN-targeting intron that cooperates with its host
gene MCM7 in transformation. Sci Signal. 3:ra292010.PubMed/NCBI
|
|
30
|
Petrocca F, Visone R, Onelli MR, Shah MH,
Nicoloso MS, de Martino I, Iliopoulos D, Pilozzi E, Liu CG, Negrini
M, et al: E2F1-Regulated MicroRNAs impair TGFbeta-dependent
cell-cycle arrest and apoptosis in gastric cancer. Cancer Cell.
13:272–286. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Han S, Park K, Bae BN, Kim KH, Kim HJ, Kim
YD and Kim HY: E2F1 expression is related with the poor survival of
lymph node-positive breast cancer patients treated with
fluorouracil, doxorubicin and cyclophosphamide. Breast Cancer Res
Treat. 82:11–16. 2003. View Article : Google Scholar : PubMed/NCBI
|