Introduction
Colorectal cancer (CRC) is the third most common
type of cancer among males and the second most common type among
females worldwide, accounting for 746,000 and 614,000 cases in
2012, respectively (1). The incidence
of CRC is rapidly increasing in developing countries, including
Malaysia, where it ranks as the second most common cancer in men
and women, with a total of 2,246 cases diagnosed in 2007 (2,3). CRC
occurs as a more frequent sporadic form and a less frequent
familial form. Familial aggregation of CRC cases results from the
inheritance of germline mutations of mismatch repair (MMR) genes.
Previously, from a cohort of CRC patients, we identified 68
suspected Lynch syndrome cases. However, an analysis of germline
mutations of the MMR genes, MLH1, MSH2, MSH6
and PMS2, confirmed only 32% of these CRC cases as Lynch
syndrome cases (4). This indicated
the existence of additional genetic susceptibility factors that
account for familial risk and which remain to be elucidated. It was
hypothesized that MMR-non-mutated familial aggregation may be
largely ‘polygenic’ due to single nucleotide polymorphisms (SNPs)
of low penetrance genes involved in cancer predisposition pathways,
including cell cycle regulation and apoptosis.
Cyclin D1 protein, encoded by CCND1, is a key
regulator of the cell cycle which modulates the transition from G1
to S phase via cyclin-dependent kinases (CDKs) during cell division
(5). However, the overexpression of
cyclin D1 due to genetic variation may disrupt cell cycle control
and potentially induce the development of cancers (6). A common polymorphism of CCND1,
G870A, has been widely investigated in numerous case-control
studies for its association with different types of cancer,
including sporadic CRC, familial CRC, squamous cell carcinoma of
the head and neck, urinary tract bladder cancer and prostate
cancer, in various populations (7–10).
Tumor protein p53, encoded by the TP53 gene,
plays a major role in regulating cell cycle progression, DNA
repair, cellular growth and apoptosis (11,12). As an
important tumor suppressor gene, the encoded protein acts to
suppress tumorigenesis and control the cell cycle checkpoint and
apoptosis under physiological stress. A common variation in
TP53, the C to G substitution at codon 72, has been
demonstrated to alter the normal function of p53. The change of
amino acid from arginine to proline in a proline-rich region of the
protein may affect the role of p53 in apoptosis (13). To date, a small number of case-control
studies have been conducted to investigate the potential
association of the CCND1 G870A and TP53 C215G
polymorphisms with CRC susceptibility risk in various populations
(14,15). However, no reports are available from
Malaysian populations. Therefore, these two SNPs were selected as
candidates for investigation, and the current case-control study
was undertaken to investigate the genotype frequencies of
CCND1 G870A and TP53 C215G polymorphisms and
determine their role in modulating familial and sporadic CRC
susceptibility risk in Malaysian subjects.
Materials and methods
Study subjects
Research conducted at the Malaysian Ministry of
Health hospitals in the present study was approved by the Research
and Ethics Committee of the School of Medical Sciences, University
of Science Malaysia (USM; Kubang Kerian, Malaysia) and the National
Institutes of Health (registration ID: NMRR-08-711-1866). Subjects
were recruited from various hospitals in Malaysia, including
Hospital USM, Hospital Sultanah Bahiyah (Alor Setar, Malaysia),
Hospital Raja Perempuan Zainab II (Kota Bharu, Malaysia) and
Hospital Queen Elizabeth (Kota Kinabalu, Malaysia) between March 1,
2008 and February 28, 2011. This case-control study involved 208
study subjects, comprising 52 histopathologically confirmed
sporadic CRC patients, 52 familial CRC patients and 104 healthy
normal controls. Initially, 68 families with suspected Lynch
syndrome [also known as hereditary nonpolyposis colorectal cancer
(HNPCC)] were identified based on Bethesda Guidelines (16): i) CRC and age <50 years; ii)
presence of synchronous or metachronous colorectal or other
HNPCC-associated tumors, regardless of age; iii) CRC with
microsatellite instability-positive morphology and age <60
years; iv) CRC and one or more first-degree relatives with CRC or
other HNPCC-related tumors, with one of the cancers occurring
<50 years of age; or v) CRC and two or more first- or
second-degree relatives with CRC or other HNPCC-related tumor
(regardless of age), including endometrial, stomach, ovarian,
cervical, esophageal, leukemia, thyroid, bladder, ureter and renal
pelvis, biliary tract, small bowel, breast, pancreas, liver,
larynx, bronchus, lung and brain cancers (glioblastoma), sebaceous
gland adenomas and keratoacanthomas. Personal and demographic
details of the patients, including family history of CRC, were
collected and recorded. CRC patients with a strong family history
of CRC among first- or second-degree relatives were subjected to a
detailed pedigree analysis. Pedigrees of the 68 suspected Lynch
syndrome families were prepared, and these patients were subjected
to protein expression and germline mutation analysis of MMR genes.
Of these, 16 cases in which germline mutations of any of the 4 MMR
genes, MLH1, MSH2, MSH6 or PMS2, were
identified were excluded from the present study, and the remaining
52 cases were included as familial CRC cases. Cases with known
familial adenomatous polyposis, ulcerative colitis or Crohn's
disease, or any other previous malignancy as stated in the
pathology reports were also excluded. For comparison, 52
histopathologically confirmed sporadic colorectal cancer patients
and 104 normal controls were also included in the present study.
Controls were normal healthy individuals who visited Hospital USM
for problems unrelated to cancer and were aged 30–65 years. The
control subjects were biologically unrelated to the patients and
were cancer-free. Epidemiological data was collected from patients
using a pre-structured questionnaire comprising questions on
socio-demographic status, physical status, dietary factors,
occupation, tobacco/alcohol habits, previous illness and radiation
exposure.
Genotyping of CCND1 G870A and TP53
C215G polymorphisms
Blood samples (3 ml) were collected from study
participants after obtaining informed consent. Genomic DNA was
extracted from blood samples using a QIAamp DNA Blood Mini kit
(Qiagen, Hilden, Germany). Genotyping of CCND1 G870A and TP53 C215G
polymorphisms was conducted using polymerase chain reaction
(PCR)-restriction fragment length polymorphism. Regions in CCND1
and TP53 containing the polymorphic site were amplified using the
following primers: CCND1 G870A forward,
5′-GTTATGTTTGAGTCAACAGTGG-3′, and reverse,
5′-TCTAGGAGCAGTGGAAGAAG-3′ (product length, 574 bp); and TP53
forward, 5′-TCAAACATCCTGTCCCTACT-3′, and reverse,
5′-CTGCGATTAAAGGCTGTGGA-3′ (product length, 458 bp). PCR was
performed in a final volume of 25 µl, consisting of 2 mM
MgCl2, 1X GeneAmp PCR Buffer II, 0.2 mM dNTPs, 0.4 µM of
each forward and reverse specific primers, 4 ng/µl of template DNA
and 1 U of AmpliTaq Gold DNA Polymerase (reagents purchased from
Applied Biosystems Life Technologies, Foster City, CA, USA). PCR
was conducted in an Eppendorf Mastercycler Gradient (Eppendorf,
Hamburg, Germany) and the conditions were as follows:
Pre-denaturation at 94°C for 5 min; 35 cycles of denaturation at
94°C for 30 sec, annealing at 57°C (CCND1 G870A) or 56°C (TP53
C215G) for 30 sec, and extension at 72°C for 30 sec; and a 72°C
final extension step for 5 min. Amplicons were then detected by gel
electrophoresis on a 2% agarose gel. Following amplification, PCR
products containing the CCND1 G870A or TP53 C215G polymorphic sites
were digested using BsrI and BstUI restriction enzymes (New England
Biolabs Inc., Ipswich, MA, USA) at 65°C for 1 h or 60°C for 1 h,
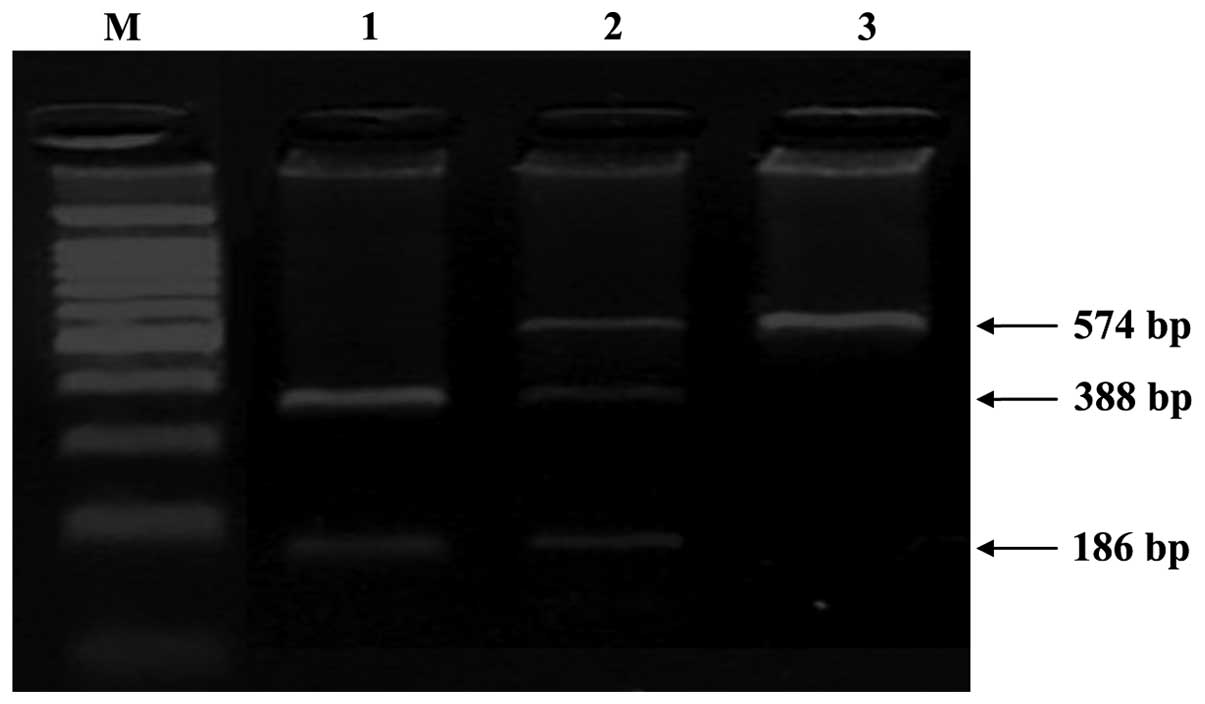
respectively. For CCND1 G870A, the G allele was not cleaved by BsrI
and thus yielded a single fragment (574 bp), whereas the A allele
was cleaved and therefore produced two fragments (388 and 186 bp);
the heterozygous genotype yielded three fragments (574, 388 and 186
bp). Genotypes were categorized as homozygous wild type (G/G),
heterozygous (G/A) or homozygous variant (A/A) based on the
fragment sizes, as shown in Fig. 1.
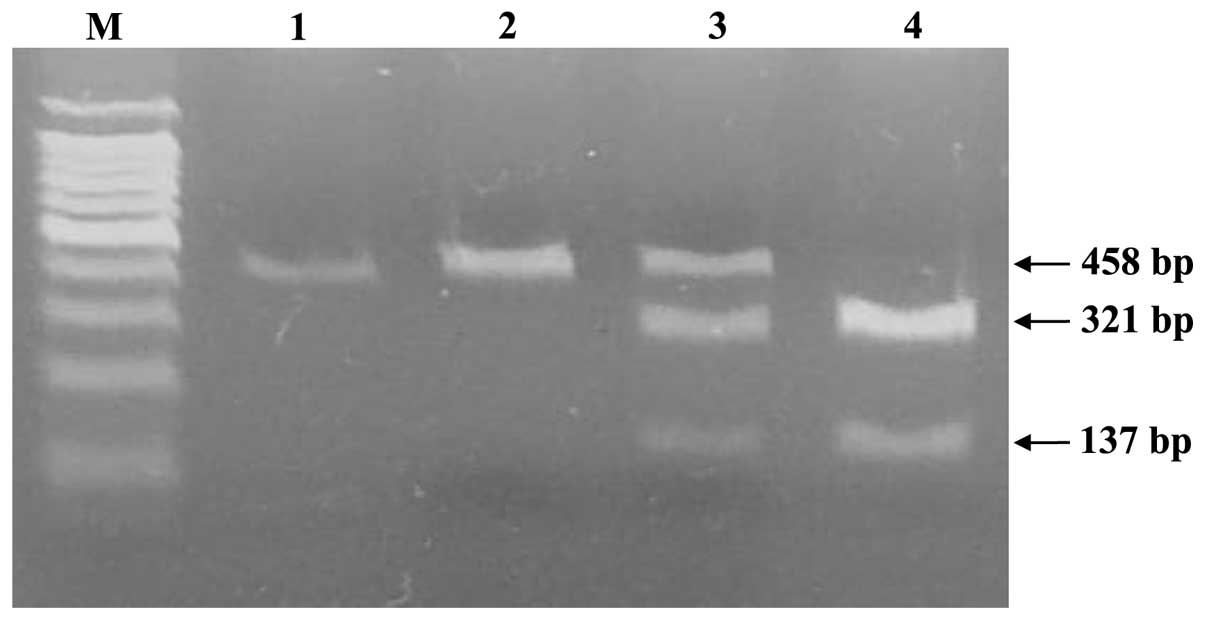
For the TP53 C215G polymorphism, the C allele was cleaved by BstUI
and thus yielded two fragments (321 and 137 bp) whereas the G
allele was not cleaved and produced a single fragment (458 bp), and
the heterozygous genotype yielded three fragments (458, 321 and 137
bp). The genotype was categorized as homozygous wild type (C/C),
heterozygous (C/G) and homozygous variant (G/G) based on the
fragment sizes (Fig. 2).
Statistical analysis
The χ2 test was used to compare the
frequency distribution of CCND1 G870A and TP53 C215G genotypes in
sporadic CRC patients, familial CRC patients and controls. The odds
ratios (ORs) and 95% confidence intervals (CIs) were calculated
using a binary logistic regression method (SPSS software version
18; SPSS, Inc., Chicago, IL, USA) to assess the risk association
between CCND1 G870A and TP53 C215G polymorphisms and sporadic CRC
and familial CRC. All statistical tests were two sided and
P<0.05 was considered to indicate statistical significance.
Results
A total of 104 CRC patients (52 sporadic CRC and 52
familial CRC patients) and 104 normal controls were recruited. Of
the 52 sporadic CRC patients, 28 were male and 24 were female, with
a mean age (± standard deviation) of 60.31±11.29 years. The 52
familial CRC patients comprised 30 males and 22 females, with a
mean age of 45.62±10.60 years. The normal controls comprised of 50
males and 54 females with a mean age of 49.62±10.78 years (Table I). The frequency distribution of SNP
genotypes was evaluated after genotyping the 208 study subjects.
The genotype frequencies of CCND1 G870A and TP53
C215G polymorphisms in CRC patients (sporadic CRC and familial CRC
cases together and separately) and normal controls are shown in
Table II.
 | Table I.Distribution of gender and age among
colorectal cancer cases and controls. |
Table I.
Distribution of gender and age among
colorectal cancer cases and controls.
|
| Colorectal
cancer |
|
|---|
|
|
|
|
|---|
| Variable | Sporadic
(n=52) | Familial
(n=52) | Control
(n=104) |
|---|
| Gender, n |
|
|
|
|
Male | 28 | 30 | 50 |
|
Female | 24 | 22 | 54 |
| Age, years |
|
Range | 36–83 | 26–72 | 27–72 |
| Mean ±
SD | 60.31±11.29 | 45.62±10.60 | 49.62±10.78 |
 | Table II.Genotype frequencies of CCND1
G870A and TP53 C215G polymorphisms in colorectal cancer
cases and normal controls. |
Table II.
Genotype frequencies of CCND1
G870A and TP53 C215G polymorphisms in colorectal cancer
cases and normal controls.
|
|
| Total cases | Sporadic CRC | Familial CRC |
|
|---|
|
|
|
|
|
|
|
|---|
| Polymorphism | Controls, n
(%) | n (%) | P-value | n (%) | P-value | n (%) | P-value |
|---|
| CCND1
G870A |
|
|
|
|
|
|
|
|
G/G | 36
(34.6%) | 20
(19.2%) | 0.017a | 10 (19.2%) | 0.047a | 10 (19.2%) | 0.047a |
|
G/A | 40
(38.5%) | 33
(31.7%) | 0.309 | 15 (28.8%) | 0.236 | 18 (34.6%) | 0.640 |
|
A/A | 28
(26.9%) | 51
(49.1%) | 0.001a | 27 (52.0%) | 0.002a | 24 (46.2%) | 0.016a |
| TP53
C215G |
|
|
|
|
|
|
|
|
C/C | 33
(31.7%) | 27
(26.0%) | 0.358 | 15 (28.9%) | 0.713 | 12 (23.1%) | 0.261 |
|
C/G | 57
(54.8%) | 43
(41.3%) | 0.052 | 19 (36.5%) | 0.031a | 24 (46.2%) | 0.308 |
|
G/G | 14
(13.5%) | 34
(32.7%) | 0.001a | 18 (34.6%) | 0.002a | 16 (30.7%) | 0.010a |
| Total | 104 (100%) | 104 (100%) |
| 52 (100%) |
| 52 (100%) |
|
The association of CCND1 G870A and
TP53 C215G polymorphisms with CRC susceptibility risk was
analyzed using a binary logistic regression analysis. ORs were
calculated relative to subjects, with the wild type G/G genotype of
CCND1 and the C/C genotype of TP53 used as
references. The A/A variant genotype of CCND1 (OR, 3.471;
CI, 1.443–8.350; P=0.005) and G/G variant genotype of TP53
(OR, 2.829; CI, 1.119–7.152; P=0.026) were revealed to be
significantly associated with the risk of sporadic CRC (Table III). Notably, these variant
genotypes of CCND1 (A/A) and TP53 (G/G) also
exhibited a significantly greater association with the risk of
familial CRC relative to the wild type genotypes (CCND1: OR,
3.086; CI, 1.270–7.497; P=0.019. TP53: OR, 3.048; CI,
1.147–8.097; P=0.030) (Table
IV).
 | Table III.Association of CCND1 G870A and
TP53 C215G polymorphisms with sporadic CRC susceptibility
risk. |
Table III.
Association of CCND1 G870A and
TP53 C215G polymorphisms with sporadic CRC susceptibility
risk.
| Polymorphism | Sporadic CRC,
n | Controls, n | OR (95% CI) | P-value |
|---|
| CCND1
G870A |
|
|
|
|
|
G/G | 10 | 36 | 1.000
(Ref.)a | – |
|
G/A | 15 | 40 | 1.350
(0.539–3.381) | 0.522 |
|
A/A | 27 | 28 | 3.471
(1.443–8.350) | 0.005b |
| TP53
C215G |
|
|
|
|
|
C/C | 15 | 33 | 1.000
(Ref.)a | – |
|
C/G | 19 | 57 | 0.733
(0.329–1.634) | 0.446 |
|
G/G | 18 | 14 | 2.829
(1.119–7.152) | 0.026b |
| Total | 52 | 104 |
|
|
 | Table IV.Association of CCND1 G870A and
TP53 C215G polymorphisms with familial CRC susceptibility
risk. |
Table IV.
Association of CCND1 G870A and
TP53 C215G polymorphisms with familial CRC susceptibility
risk.
| Polymorphism | Familial CRC,
n | Controls, n | OR (95% CI) | P-value |
|---|
| CCND1
G870A |
|
|
|
|
|
G/G | 10 | 36 | 1.000
(Ref.)a | – |
|
G/A | 18 | 40 | 1.620
(0.662–3.963) | 0.374 |
|
A/A | 24 | 28 | 3.086
(1.270–7.497) | 0.019b |
| TP53
C215G |
|
|
|
|
|
C/C | 12 | 33 | 1.000
(Ref.)a | – |
|
C/G | 24 | 57 | 1.103
(0.488–2.496) | 0.806 |
|
G/G | 16 | 14 | 3.048
(1.147–8.097) | 0.030b |
| Total | 52 | 104 |
|
|
Discussion
The incidence of CRC has been increasing rapidly,
and the disease has become one of the leading causes of
cancer-related mortality worldwide in recent years. As a result,
large numbers of research studies have been directed towards the
investigation of CRC, particularly with regard to its etiology,
prevention and treatment (17–19).
Colorectal carcinogenesis is a complex, gradual and
multistep process involving various factors (20,21). The
protein encoded by CCND1, cyclin D1, is involved in
colorectal carcinogenesis in its role as one of the key regulatory
proteins of the cell cycle transition from G1 to S phase (5,22).
CCND1 G870A (Pro241Pro) is a silent variant in which the
alteration of amino acids does not occur. However, as this
variation is in the last nucleotide of exon 4, it may lead to the
alternative splicing of CCND1 mRNA. Both alleles are capable
of producing two different transcripts, a normal splicing exon 4
and exon 5, known as ‘transcript a’, and an alternative transcript,
‘transcript b’, lacking exon 5, the exon comprising a PEST domain
responsible for the degradation of cyclin D1 protein (23,24).
Therefore, it has been reported that the variant A allele may
produce a cyclin D1 protein with an increased half life, and the
resulting accumulation of the protein may promote cell
proliferation (25).
According to Fearon and Vogelstein (26), colorectal carcinogenesis involves a
number of genetic changes that include mutation of certain critical
genes, such as TP53, and these mutations accumulate during
the progression from normal epithelium to carcinoma. TP53 is
one of the most extensively studied genes due to its crucial role
in regulating the cell cycle, apoptosis, inhibition of angiogenesis
and cellular senescence (27).
Mutations or variations in TP53 may completely diminish the
function of the p53 protein, and thus may promote cell
proliferation and carcinogenesis. The amino acid substitution
caused by the C215G SNP may alter the conformation of the protein
and, therefore, its ability to bind to elements in target genes,
and may affect the stability and interaction of the protein with
the other proteins (28,29). Previous studies have described
functional differences that are caused by these two alleles in
TP53. For example, Pim and Banks (30) reported that the wild type G (Arg72)
allele is more efficient in promoting apoptosis compared with the
variant C (Pro72) allele. A similar finding also has been observed
by Marin et al (31), who
reported that the mutant p53 protein exhibited a reduced efficacy
during apoptosis compared with the Arg allele protein. Thus, these
studies provide evidence that variation in TP53 may lead to
abnormal protein function and contribute to cell proliferation and
cancer.
The present study investigated the genotype
frequencies of CCND1 G870A and TP53 C215G
polymorphisms and the potential association with susceptibility
risk in Malaysian CRC patients. The results revealed that the
variant genotypes of CCND1 (A/A) and TP53 (G/G) have
a significantly higher risk association with sporadic CRC relative
to the homozygous wild type genotypes (OR, 3.471 and 2.829,
respectively; P<0.05; Table
III). In addition, the variant genotypes of the two SNPs
exhibited a significantly higher risk association with familial CRC
compared with the wild type genotypes (OR, 3.086 and 3.048,
respectively; P<0.05; Table IV).
The results suggest that individuals with the A/A genotype of
CCND1 G870A and G/G genotype of TP53 C215G have a
three-fold higher risk for CRC development compared with
individuals possessing the G/G and C/C wild type genotypes.
The association of the CCND1 G870A
polymorphism with CRC susceptibility risk has been reported
previously in a number of studies (32–34). The
present results are in concordance with studies conducted by Hong
et al (14) and Jiang et
al (35), in which the A/A
genotype of CCND1 G870A was found to be associated with CRC
susceptibility risk in Singaporean and Indian patients (OR, 2.4 and
1.56, respectively). Another Indian case-control study conducted in
a population of Kashmiri ethnicity also identified an association
between the variant genotype of CCND1 G870A and risk of CRC,
with a two-fold increase in OR relative to the wild type genotype
(34). Previous meta-analyses
involving numerous case-control studies also demonstrated an
association between CCND1 G870A polymorphism and CRC
susceptibility risk (32,33), suggesting that the CCND1 870A
allele may be a low-penetrant risk factor for CRC.
On the other hand, conflicting results have been
reported on the possible association between TP53 C215G
polymorphisms and CRC susceptibility. The G/G variant genotype was
found to be associated with an increased risk of CRC in Japanese,
Korean and Indian Kashmiri populations (36–38). A
case-control study involving 444 sporadic CRC patients in China
also reported that the G/G variant genotype was associated with an
increased risk of CRC (39). In
addition, an earlier study conducted by our group (40) revealed that the G/G genotype had a
significant risk association with sporadic CRC susceptibility in a
Malaysian population (OR, 2.047; CI, 1.063–4.044; P=0.013). By
contrast, meta-analyses conducted by Economopoulos et al
(41) and Dahabreh et al
(42) did not identify any
significant risk association between G/G genotype and CRC
susceptibility. Furthermore, Oh et al (43) reported that the variant genotype of
TP53 C215G was associated with a decreased risk of CRC.
Notably, in addition to their association with
sporadic CRC, the variant genotypes of both SNPs investigated in
the present study (A/A in CCND1 and G/G in TP53),
exhibited a significant association with the risk of familial CRC
(Table IV). However, few other
studies have been conducted on the association of these SNPs with
familial CRC susceptibility risk, and the results have been
contradictory. In a study by Chen et al (44), CCND1 G870A was demonstrated to
contribute to the early onset of CRC in cases of Lynch syndrome,
whilst the TP53 C215G SNP was not. In a meta-analysis
involving 20 different populations, the variant allele of
CCND1 G870A was found to be significantly associated with an
elevated CRC risk (OR, 1.23); however, no association with CRC risk
was observed in Lynch syndrome patients (32). Rather than focusing on CRC
susceptibility risk, the majority of previous studies have been
focusing on the impact of CCND1 G870A and TP53 C215G
on the age of onset in familial CRC cases, including cases of Lynch
syndrome (45,46). Krüger et al (47) observed no association between
CCND1 G870A and age of onset of Lynch syndrome among 406 MMR
mutation carriers. Conflicting results also have been reported
regarding the association of TP53 C215G polymorphism with
the risk of CRC in Lynch syndrome patients. In other studies, the
age of diagnosis of CRC in Lynch syndrome patients was not
associated with TP53 C215G genotype (46,48). In
the present study, CRC patients identified to harbor MMR mutations
were excluded, and the association of the two SNPs with age of
onset of CRC in Lynch syndrome patients was therefore not
investigated.
In summary, our results provide evidence of the
effect of CCND1 G870A and TP53 C215G polymorphisms on
CRC susceptibility risk. Despite the small sample size, the two
SNPs have been observed to contribute to the susceptibility risk of
sporadic and familial CRC in a Malaysian population. Individuals
with CCND1 G870A A/A and TP53 C215G G/G genotypes in
particular may have a higher risk for sporadic and familial CRC
susceptibility. However, further studies into polymorphisms of
other genes involved in pathways associated with sporadic and
familial CRC in larger sample size, in addition to functional
studies, are required to confirm these findings.
Acknowledgements
The authors would like to thank Dato' Dr Muhammad
Radzi Abu Hassan (Hospital Sultanah Bahiyah) and Dr Ahmad Shanwani
Mohd Sidek (Hospital Raja Perempuan Zainab II) for their assistance
in recruiting the study subjects. The authors also wish to thank
all the study participants for their contribution. This study was
supported by a USM Research University grant (no.
1001/PPSP/812070).
References
|
1
|
Ferlay J, Soerjomataram I, Ervik M,
Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D and
Bray F: GLOBOCAN 2012 v1.0. Cancer Incidence and Mortality
Worldwide: IARC CancerBase No. 11 (Internet). International Agency
for Research on Cancer (Lyon, France). 2013.Available from.
http://globocan.iarc.frAccessed.
July 27–2015
|
|
2
|
Goh KL, Quek KF, Yeo GT, Hilmi IN, Lee CK,
Hasnida N, Aznan M, Kwan KL and Ong KT: Colorectal cancer in
Asians: A demographic and anatomic survey in Malaysian patients
undergoing colonoscopy. Aliment Pharmacol Ther. 22:859–864. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
National Cancer Registry: Malaysia Cancer
Statistics - Data and Figure: Peninsular Malaysia 2006. Kuala
Lumpur: National Cancer Registry, Ministry of Health Malaysia.
2006.
|
|
4
|
Zahary MN, Kaur G, Hassan MR, Sidek AS,
Singh H, Yeh LY and Ankathil R: Germline mutation and protein
expression analysis of mismatch repair genes MSH6 and PMS2 in
Malaysian Lynch syndrome patients. Int J Colorectal Dis.
29:261–262. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Sherr CJ: Cancer cell cycles. Science.
274:1672–1677. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Donnellan R and Chetty R: Cyclin D1 and
human neoplasia. Mol Pathol. 51:1–7. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zheng Y, Shen H, Sturgis EM, Wang LE,
Eicher SA, Strom SS, Frazier ML, Spitz MR and Wei Q: Cyclin D1
polymorphism and risk for squamous cell carcinoma of the head and
neck: A case-control study. Carcinogenesis. 22:1195–1199. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Porter TR, Richards FM, Houlston RS, Evans
DG, Jankowski JA, Macdonald F, Norbury G, Payne SJ, Fisher SA,
Tomlinson I and Maher ER: Contribution of cyclin d1 (CCND1) and
E-cadherin (CDH1) polymorphisms to familial and sporadic colorectal
cancer. Oncogene. 21:1928–1933. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wang L, Habuchi T, Takahashi T, Mitsumori
K, Kamoto T, Kakehi Y, Kakinuma H, Sato K, Nakamura A, Ogawa O and
Kato T: Cyclin D1 gene polymorphism is associated with an increased
risk of urinary bladder cancer. Carcinogenesis. 23:257–264. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wang L, Habuchi T, Mitsumori K, Li Z,
Kamoto T, Kinoshita H, Tsuchiya N, Sato K, Ohyama C, Nakamura A, et
al: Increased risk of prostate cancer associated with AA genotype
of cyclin D1 gene A870G polymorphism. Int J Cancer. 103:116–120.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lane DP: Cancer. p53, guardian of the
genome. Nature. 358:15–16. 1992. View
Article : Google Scholar : PubMed/NCBI
|
|
12
|
Levine AJ: p53, the cellular gatekeeper
for growth and division. Cell. 88:323–331. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Sullivan A, Syed N, Gasco M, Bergamaschi
D, Trigiante G, Attard M, Hiller L, Farrell PJ, Smith P, Lu X and
Crook T: Polymorphism in wild-type p53 modulates response to
chemotherapy in vitro and in vivo. Oncogene. 23:3328–3337. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hong Y, Eu KW, Seow-Choen F, Fook-Chong S
and Cheah PY: GG genotype of cyclin D1 G870A polymorphism is
associated with increased risk and advanced colorectal cancer in
patients in Singapore. Eur J Cancer. 41:1037–1044. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Cao Z, Song JH, Park YK, Maeng EJ, Nam SW,
Lee JY and Park WS: The p53 codon 72 polymorphism and
susceptibility to colorectal cancer in Korean patients. Neoplasma.
56:114–118. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Umar A, Boland CR, Terdiman JP, Syngal S,
de la Chapelle A, Rüschoff J, Fishel R, Lindor NM, Burgart LJ,
Hamelin R, et al: Revised Bethesda Guidelines for hereditary
nonpolyposis colorectal cancer (Lynch syndrome) and microsatellite
instability. J Natl Cancer Inst. 96:261–268. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Tárraga López PJ, Albero JS and
Rodríguez-Montes JA: Primary and secondary prevention of colorectal
cancer. Clin Med Insights Gastroenterol. 7:33–46. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Marin JJ, de Medina Sanchez F, Castaño B,
Bujanda L, Romero MR, Martinez-Augustin O, Moral-Avila RD and Briz
O: Chemoprevention, chemotherapy and chemoresistance in colorectal
cancer. Drug Metab Rev. 44:148–172. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Sengupta N, Gill KA, MacFie TS, Lai CS,
Suraweera N, Mcdonald S and Silver A: Management of colorectal
cancer: A role for genetics in prevention and treatment? Pathol Res
Pract. 204:469–477. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Gryfe R, Swallow C, Bapat B, Redston M,
Gallinger S and Couture J: Molecular biology of colorectal cancer.
Curr Probl Cancer. 21:233–300. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Bellacosa A: Genetic hits and mutation
rate in colorectal tumorigenesis: Versatility of Knudson's theory
and implications for cancer prevention. Genes Chromosomes Cancer.
38:382–388. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Shtutman M, Zhurinsky J, Simcha I,
Albanese C, D'Amico M, Pestell R and Ben-Ze'ev A: The cyclin D1
gene is a target of the beta-catenin/LEF-1 pathway. Proc Natl Acad
Sci U S A. 96:5522–5527. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Betticher DC, Thatcher N, Altermatt HJ,
Hoban P, Ryder WD and Heighway J: Alternate splicing produces a
novel cyclin D1 transcript. Oncogene. 11:1005–1011. 1995.PubMed/NCBI
|
|
24
|
Solomon DA, Wang Y, Fox SR, Lambeck TC,
Giesting S, Lan Z, Senderowicz AM, Conti CJ and Knudsen ES: Cyclin
D1 splice variants. Differential effects on localization, RB
phosphorylation and cellular transformation. J Biol Chem.
278:30339–30347. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Sawa H, Ohshima TA, Ukita H, Murakami H,
Chiba Y, Kamada H, Hara M and Saito I: Alternatively spliced forms
of cyclin D1 modulate entry into the cell cycle in an inverse
manner. Oncogene. 16:1701–1712. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Fearon ER and Vogelstein B: A genetic
model for colorectal tumorigenesis. Cell. 61:759–767. 1990.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Whibley C, Pharoah PD and Hollstein M: p53
polymorphisms: Cancer implications. Nat Rev Cancer. 9:95–107. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Thomas M, Kalita A, Labrecque S, Pim D,
Banks L and Matlashewski G: Two polymorphic variants of wild-type
p53 differ biochemically and biologically. Mol Cell Biol.
19:1092–1100. 1999.PubMed/NCBI
|
|
29
|
Bergamaschi D, Gasco M, Hiller L, Sullivan
A, Syed N, Trigiante G, Yulug I, Merlano M, Numico G, Comino A, et
al: p53 polymorphism influences response in cancer chemotherapy via
modulation of p73-dependent apoptosis. Cancer Cell. 3:387–402.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Pim D and Banks L: p53 polymorphic
variants at codon 72 exert different effects on cell cycle
progression. Int J Cancer. 108:196–199. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Marin MC, Jost CA, Brooks LA, Irwin MS,
O'Nions J, Tidy JA, James N, McGregor JM, Harwood CA, Yulug IG, et
al: A common polymorphism acts as an intragenic modifier of mutant
p53 behaviour. Nat Genet. 25:47–54. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zhang LQ, Wang J, Shang JQ, Bai JL, Liu
FY, Guan X and Zhou JN: Cyclin D1 G870A polymorphism and colorectal
cancer susceptibility: A meta-analysis of 20 populations. Int J
Colorectal Dis. 26:1249–1255. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Yang Y, Wang F, Shi C, Zou Y, Qin H and Ma
Y: Cyclin D1 G870A polymorphism contributes to colorectal cancer
susceptibility: Evidence from a sytematic review of 22 case-control
studies. PLoS One. 7:e368132012. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Sameer AS, Parray FQ, Dar MA, Nissar S,
Banday MZ, Rasool S, Gulzar GM, Chowdri NA and Siddiqi MA: Cyclin
D1 G870A polymorphism and risk of colorectal cancer: A case control
study. Mol Med Rep. 7:811–815. 2013.PubMed/NCBI
|
|
35
|
Jiang J, Wang J, Suzuki S, Gajalakshmi V,
Kuriki K, Zhao Y, Nakamura S, Akasaka S, Ishikawa H and Tokudome S:
Elevated risk of colorectal cancer associated with the AA genotype
of the cyclin D1 A870G polymorphism in an Indian population. J
Cancer Res Clin Oncol. 132:193–199. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Sameer AS, Shah ZA, Syeed N, Banday MZ,
Bashir SM, Bhat BA and Siddiqi MA: TP53 Pro47Ser and Arg72Pro
polymorphisms and colorectal cancer predisposition in an ethnic
Kashmiri population. Genet Mol Res. 9:651–660. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Joshi AM, Budhathoki S, Ohnaka K, Mibu R,
Tanaka M, Kakeji Y, Maehara Y, Okamura T, Ikejiri K, Futami K, et
al: TP53 R72P and MDM2 SNP309 polymorphisms and colorectal cancer
risk: The Fukuoka Colorectal Cancer Study. Jpn J Clin Oncol.
41:232–238. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Song HR, Kweon SS, Kim HN, Piao JM, Yun
WJ, Choi JS, Hwang JE, Yoon JY, Kim HR, Park YK, et al: p53 codon
72 polymorphism in patients with gastric and colorectal cancer in a
Korean population. Gastric Cancer. 14:242–248. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Zhang Y, Liu L, Tang Y, et al:
Polymorphisms in TP53 and MDM2 contribute to higher risk of
colorectal cancer in Chinese population: A hospital-based, case
control study. Mol Biol Rep. 39:9661–9668. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Aizat AA, Shahpudin SN, Mustapha MA, et
al: Association of Arg72Pro of P53 polymorphism with colorectal
cancer susceptibility risk in Malaysian population. Asian Paci J
Cancer Prev. 12:2909–2913. 2011.
|
|
41
|
Economopoulos KP, Sergentanis TN, Zagouri
F and Zografos GC: Association between p53 Arg72Pro polymorphism
and colorectal cancer risk: A meta-analysis. Onkologie. 33:666–674.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Dahabreh IJ, Linardou H, Bouzika P,
Varvarigou V and Murray S: TP53 Arg72Pro polymorphism and
colorectal cancer risk: A systematic review and meta-analysis.
Cancer Epidemiol Biomarkers Prev. 19:1840–1847. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Oh J, Kim JW, Lee BE, Jang MJ, Chong SY,
Park PW, Hwang SG, Oh D and Kim NK: Polymorphisms of the
pri-miR-34b/c promoter and TP53 codon 72 are associated with risk
of colorectal cancer. Oncol Rep. 31:995–1002. 2014.PubMed/NCBI
|
|
44
|
Chen J, Etzel CJ, Amos CI, Zhang Q,
Viscofsky N, Lindor NM, Lynch PM and Frazier ML: Genetic variant in
the cell cycle control pathways contribute to early onset
colorectal cancer in Lynch syndrome. Cancer Causes Control.
20:1769–1777. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Krüger S, Bier A, Engel C, Mangold E,
Pagenstecher C, von Knebel Doeberitz M, Holinski-Feder E, Moeslein
G, Schulmann K, Plaschke J, et al: The p53 codon 72 variation is
associated with the age of onset of hereditary non-polyposis
colorectal cancer (HNPCC). J Med Genet. 42:769–773. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Sotamaa K, Liyanarachchi S, Mecklin JP,
Järvinen H, Aaltonen LA, Peltomäki P and de la Chapelle A: p53
codon 72 and MDM2 SNP309 polymorphisms and age of colorectal cancer
onset in Lynch syndrome. Clin Cancer Res. 11:6840–6844. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Krüger S, Engel C, Bier A, Mangold E,
Pagenstecher C, Doeberitz MV, Holinski-Feder E, Moeslein G, Keller
G, Kunstmann E, et al: Absence of association between cyclin D1
(CCND1) G870A polymorphism and age of onset in hereditary
nonpolyposis colorectal cancer. Cancer Lett. 236:191–197. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Talseth BA, Meldrum C, Suchy J, Kurzawski
G, Lubinski J and Scott RJ: MDM2 SNP309 T>G alone or in
combination with the TP53 R72P polymorphism does not appear to
influence disease expression and age of diagnosis of colorectal
cancer in HNPCC patients. Int J Cancer. 120:563–565. 2007.
View Article : Google Scholar : PubMed/NCBI
|