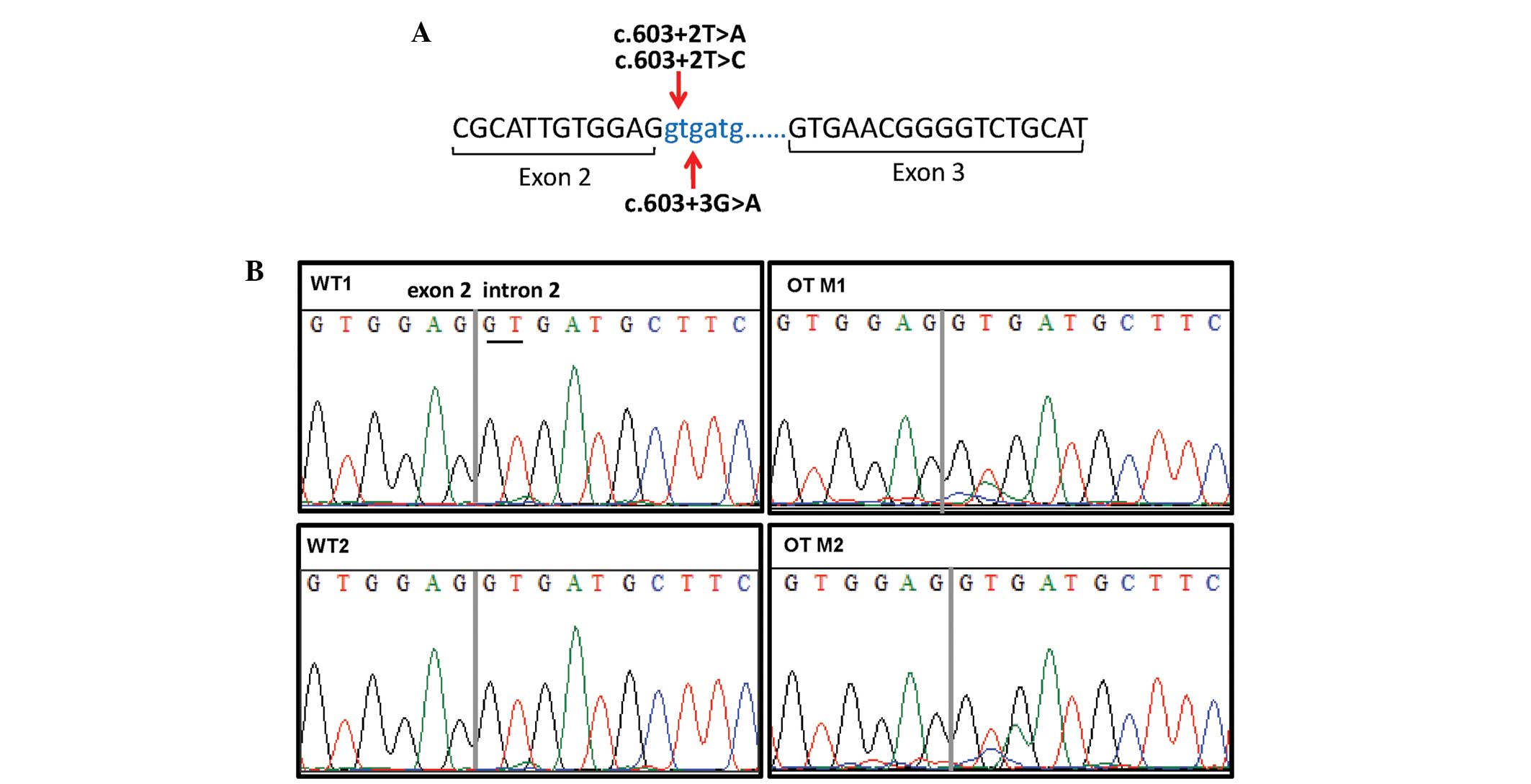
|
1
|
Ferlay J, Soerjomataram I, Ervik M,
Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D and
Bray F: GLOBOCAN 2012 v1.0, Cancer Incidence and Mortality
Worldwide: IARC CancerBase No. 11 (Internet). International Agency
for Research on Cancer (Lyon, France). 2013.http://globocan.iarc.frAccessed on. Dec
13–2013
|
|
2
|
Tinker AV, Boussioutas A and Bowtell DD:
The challenges of gene expression microarrays for the study of
human cancer. Cancer Cell. 9:333–339. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Weinman EJ, Steplock D, Wang Y and
Shenolikar S: Characterization of a protein cofactor that mediates
protein kinase A regulation of the renal brush border membrane
Na(+)/H+ exchanger. J Clin Invest. 95:2143–2149. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Reczek D, Berryman M and Bretscher A:
Identification of EBP50: A PDZ-containing phosphoprotein that
associates with members of the ezrin-radixin-moesin family. J Cell
Biol. 139:169–179. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Cho KO, Hunt CA and Kennedy MB: The rat
brain postsynaptic density fraction contains a homolog of the
Drosophila discs-large tumor suppressor protein. Neuron. 9:929–942.
1992. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Saotome I, Curto M and McClatchey AI:
Ezrin is essential for epithelial organization and villus
morphogenesis in the developing intestine. Dev Cell. 6:855–864.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Bretscher A, Chambers D, Nguyen R and
Reczek D: ERM-Merlin and EBP50 protein families in plasma membrane
organization and function. Annu Rev Cell Dev Biol. 16:113–143.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Shibata T, Chuma M, Kokubu A, Sakamoto M
and Hirohashi S: EBP50, a beta-catenin-associating protein,
enhances Wnt signaling and is over-expressed in hepatocellular
carcinoma. Hepatology. 38:178–186. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kreimann EL, Morales FC, de Orbeta-Cruz J,
Takahashi Y, Adams H, Liu TJ, McCrea PD and Georgescu MM: Cortical
stabilization of beta-catenin contributes to NHERF1/EBP50 tumor
suppressor function. Oncogene. 26:5290–5299. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Hayashi Y, Molina JR, Hamilton SR and
Georgescu MM: NHERF1/EBP50 Is a new marker in colorectal cancer.
Neoplasia. 12:1013–1022. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Thiery JP: Epithelial-mesenchymal
transitions in tumour progression. Nat Rev Cancer. 2:442–454. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Hazan RB, Qiao R, Keren R, Badano I and
Suyama K: Cadherin switch in tumor progression. Ann NY Acad Sci.
1014:155–163. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Dai JL, Wang L, Sahin AA, Broemeling LD,
Schutte M and Pan Y: NHERF1 (Na+/H+ exchanger regulatory factor)
gene mutations in human breast cancer. Oncogene. 23:8681–8687.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Cancer Genome Atlas Research Network:
Integrated genomic analyses of ovarian carcinoma. Nature.
474:609–615. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Scully RE: World Health Organization
International Histological Classification of Tumours (2nd). New
York: Springer. 1999.
|
|
16
|
Hall TA: BioEdit: A user-friendly
biological sequence alignment editor and analysis program for
Windows 95/98/NT. Nucleic Acids Symp Ser. 41:95–98. 1999.
|
|
17
|
Desmet FO, Hamroun D, Lalande M,
Collod-Béroud G, Claustres M and Béroud C: Human splicing finder:
An online bioinformatics tool to predict splicing signals. Nucleic
Acids Res. 37:e672009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Shapiro MB and Senapathy P: RNA splice
junctions of different classes of eukaryotes: Sequence statistics
and functional implications in gene expression. Nucleic Acids Res.
15:7155–7174. 1987. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Morales FC, Takahashi Y, Momin S, Adams H,
Chen X and Georgescu MM: NHERF1/EBP50 head-to-tail intramolecular
interaction masks the association with PDZ-domain ligands. Mol Cell
Biol. 27:2527–2537. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Pauler DK, Menon U, McIntosh M, Symecko
HL, Skates SJ and Jacobs IJ: Factors influencing serum CA125II
levels in healthy postmenopausal women. Cancer Epidemiol Biomarkers
Prev. 10:489–493. 2001.PubMed/NCBI
|
|
21
|
Venables JP: Aberrant and alternative
splicing in cancer. Cancer Res. 64:7647–7654. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Cooper DN and Krawczak M: Human Gene
Mutation. (Oxford, UK). Bios Scientific Publishers. 111–127.
1993.
|
|
23
|
Cáceres JF and Kornblithtt AR: Alterative
splicing: Multiple control mechanisms and involvement in human
disease. Trends Genet. 18:186–193. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Baralle D and Baralle M: Splicing in
action: Assessing disease causing sequence changes. J Med Genet.
42:737–748. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Fu XD: Towards a splicing code. Cell.
119:736–738. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Blencowe BJ: Alternative splicing: New
insights from global analyses. Cell. 126:37–47. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Brinkman BM: Splice variants as cancer
biomarkers. Clin Biochem. 37:584–594. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kalnina Z, Zayakin P, Silina K and Linē A:
Alterations of pre-mRNA splicing in cancer. Genes Chromosomes
Cancer. 42:342–357. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Nelson WJ and Nusse R: Convergence of Wnt,
beta-catenin, and cadherin pathways. Science. 303:1483–1487. 2004.
View Article : Google Scholar : PubMed/NCBI
|