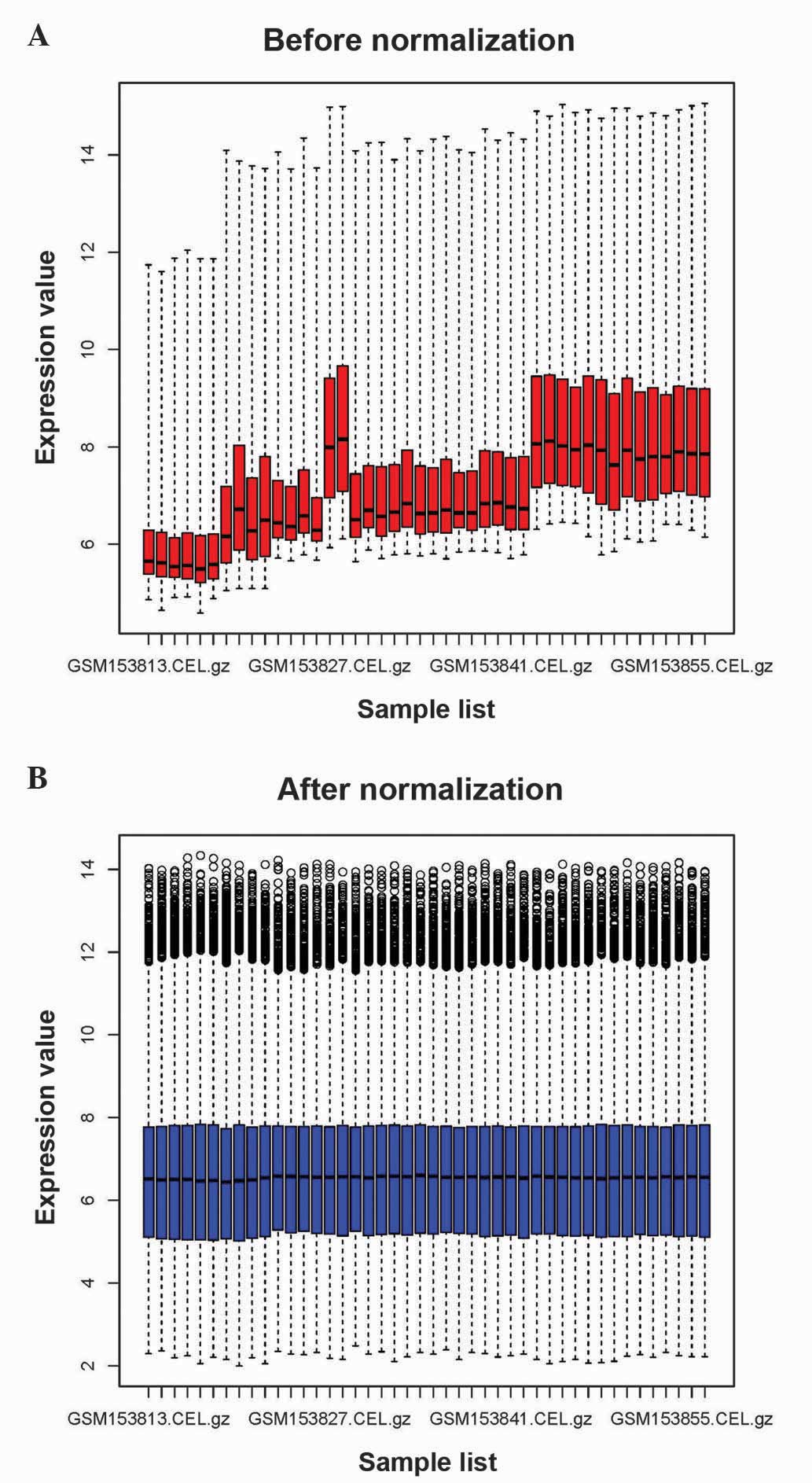
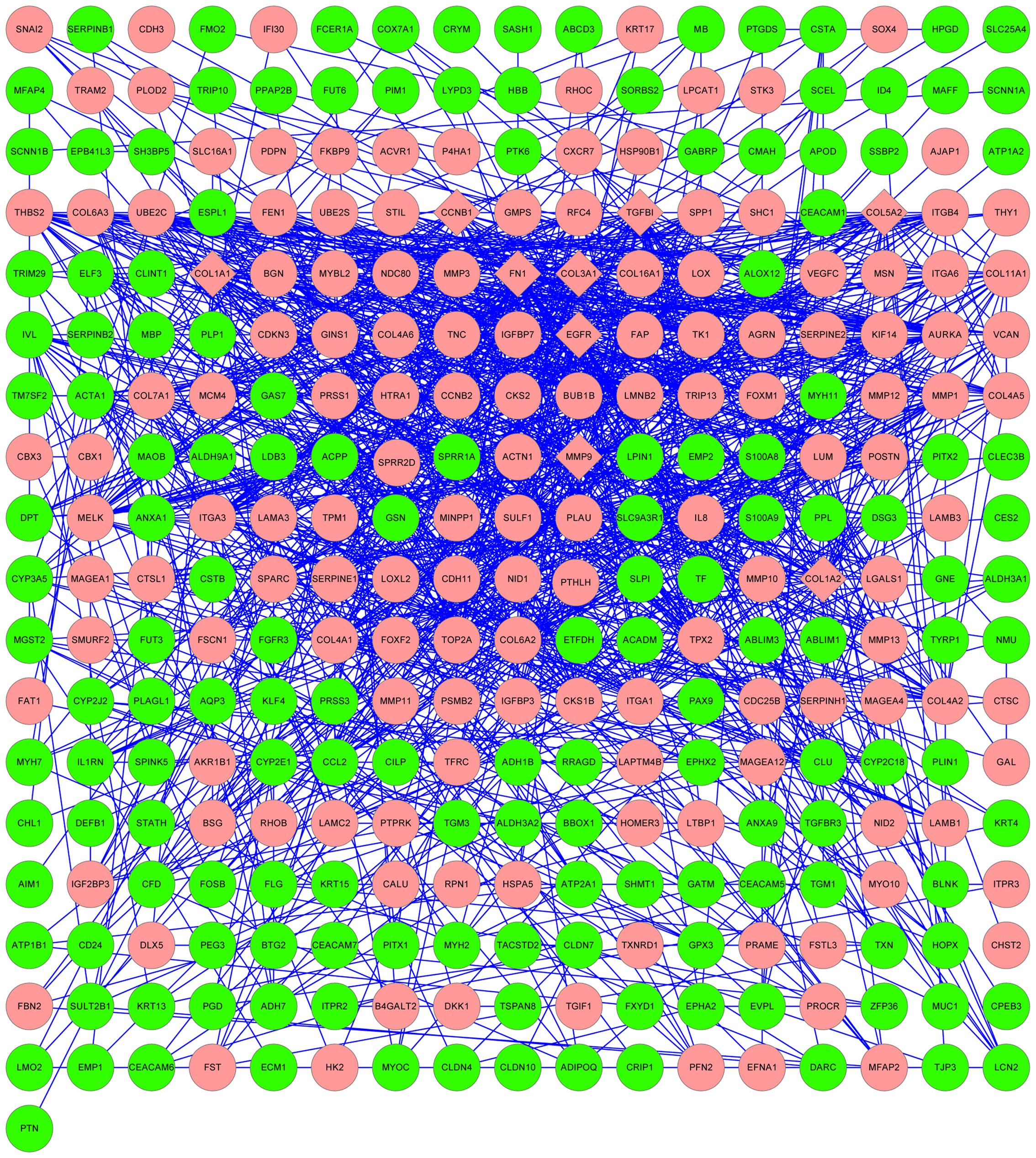
|
1
|
Ferlay J, Shin HR, Bray F, Forman D,
Mathers C and Parkin DM: Estimates of worldwide burden of cancer in
2008: GLOBOCAN 2008. Int J Cancer. 127:2893–2917. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Rothenberg SM and Ellisen LW: The
molecular pathogenesis of head and neck squamous cell carcinoma. J
Clin Invest. 122:1951–1957. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
3
|
Hasina R, Whipple ME, Martin LE, Kuo WP,
Ohno-Machado L and Lingen MW: Angiogenic heterogeneity in head and
neck squamous cell carcinoma: Biological and therapeutic
implications. Lab Invest. 88:342–353. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Belcher R, Hayes K, Fedewa S and Chen AY:
Current treatment of head and neck squamous cell cancer. J Surg
Oncol. 110:551–574. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Colevas AD: Chemotherapy options for
patients with metastatic or recurrent squamous cell carcinoma of
the head and neck. J Clin Oncol. 24:2644–2652. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Leemans CR, Braakhuis BJ and Brakenhoff
RH: The molecular biology of head and neck cancer. Nat Rev Cancer.
11:9–22. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
7
|
Nohata N, Sone Y, Hanazawa T, Fuse M,
Kikkawa N, Yoshino H, Chiyomaru T, Kawakami K, Enokida H, Nakagawa
M, et al: miR-1 as a tumor suppressive microRNA targeting TAGLN2 in
head and neck squamous cell carcinoma. Oncotarget. 2:29–42.
2011.PubMed/NCBI
|
|
8
|
Chaisaingmongkol J, Popanda O, Warta R,
Dyckhoff G, Herpel E, Geiselhart L, Claus R, Lasitschka F, Campos
B, Oakes CC, et al: Epigenetic screen of human DNA repair genes
identifies aberrant promoter methylation of NEIL1 in head and neck
squamous cell carcinoma. Oncogene. 31:5108–5116. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhang L, Pan HY, Zhong LP, Wei KJ, Yang X,
Li J, Shen GF and Zhang Z: Fos-related activator-1 is overexpressed
in oralsquamous cell carcinoma and associated with tumor lymph node
metastasis. J Oral Pathol Med. 39:470–476. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Pedrero JM, Carracedo DG, Pinto CM,
Zapatero AH, Rodrigo JP, Nieto CS and Gonzalez MV: Frequent genetic
and biochemical alterations of the PI3-K/AKT/PTEN pathway in head
and neck squamous cell carcinoma. Int J Cancer. 114:242–248. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Gallo O, Masini E, Bianchi B, Bruschini L,
Paglierani M and Franchi A: Prognostic significance of
cyclooxygenase-2 pathway and angiogenesis in head and neck squamous
cell carcinoma. Hum Pathol. 33:708–714. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Bancroft CC, Chen Z, Dong G, Sunwoo JB,
Yeh N, Park C and Van Waes C: Coexpression of proangiogenic factors
IL-8 and VEGF by human head and neck squamous cell carcinoma
involves coactivation by MEK-MAPK and IKK-NF-kappaB signal
pathways. Clin Cancer Res. 7:435–442. 2001.PubMed/NCBI
|
|
13
|
Kuriakose MA, Chen WT, He ZM, Sikora AG,
Zhang P, Zhang ZY, Qiu WL, Hsu DF, McMunn-Coffran C, Brown SM, et
al: Selection and validation of differentially expressed genes in
head and neck cancer. Cell Mol Life Sci. 61:1372–1383. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Gautier L, Cope L, Bolstad BM and Irizarry
RA: affy - analysis of Affymetrix GeneChip data at the probe level.
Bioinformatics. 20:307–315. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Tusher VG, Tibshirani R and Chu G:
Significance analysis of microarrays applied to the ionizing
radiation response. Proc Natl Acad Sci USA. 98:5116–5121. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ashburner M, Ball CA, Blake JA, Botstein
D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT,
et al: The Gene Ontology Consortium: Gene Ontology: Tool for the
unification of biology. Nat Genet. 25:25–29. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Altermann E and Klaenhammer TR:
PathwayVoyager: Pathway mapping using the Kyoto Encyclopedia of
Genes and Genomes (KEGG) database. BMC Genomics. 6:602005.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Huang DW, Sherman BT, Tan Q, Kir J, Liu D,
Bryant D, Guo Y, Stephens R, Baseler MW, Lane HC and Lempicki RA:
DAVID Bioinformatics Resources: Expanded annotation database and
novel algorithms to better extract biology from large gene lists.
Nucleic Acids Res. 35(Web Server issue): W169–W175. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Szklarczyk D, Franceschini A, Kuhn M,
Simonovic M, Roth A, Minguez P, Doerks T, Stark M, Muller J, Bork
P, et al: The STRING database in 2011: Functional interaction
networks of proteins, globally integrated and scored. Nucleic Acids
Res. 39(Database issue): D561–D568. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Smoot ME, Ono K, Ruscheinski J, Wang PL
and Ideker T: Cytoscape 2.8: New features for data integration and
network visualization. Bioinformatics. 27:431–432. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Gumbiner BM: Cell adhesion: The molecular
basis of tissue architecture and morphogenesis. Cell. 84:345–357.
1996. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Albelda SM and Buck CA: Integrins and
other cell adhesion molecules. FASEB J. 4:2868–2880.
1990.PubMed/NCBI
|
|
23
|
Wang F, Song G, Liu M, Li X and Tang H:
miRNA-1 targets fibronectin 1 and suppresses the migration and
invasion of the HEp2 laryngeal squamous carcinoma cell line. FEBS
Lett. 585:3263–3269. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Goerner M, Seiwert TY and Sudhoff H:
Molecular targeted therapies in head and neck cancer - an update of
recent developments. Head Neck Oncol. 2:82010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kleinman HK, Martin GR and Fishman PH:
Ganglioside inhibition of fibronectin-mediated cell adhesion to
collagen. Proc Natl Acad Sci USA. 76:3367–3371. 1979. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Pankov R and Yamada KM: Fibronectin at a
glance. J Cell Sci. 115:3861–3863. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Soikkeli J, Podlasz P, Yin M, Nummela P,
Jahkola T, Virolainen S, Krogerus L, Heikkilä P, von Smitten K,
Saksela O and Hölttä E: Metastatic outgrowth encompasses COL-I,
FN1, and POSTN up-regulation and assembly to fibrillar networks
regulating cell adhesion, migration, and growth. Am J Pathol.
177:387–403. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Mao L, Hong WK and Papadimitrakopoulou VA:
Focus on head and neck cancer. Cancer Cell. 5:311–316. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Erjala K, Sundvall M, Junttila TT, Zhang
N, Savisalo M, Mali P, Kulmala J, Pulkkinen J, Grenman R and
Elenius K: Signaling via ErbB2 and ErbB3 associates with resistance
and epidermal growth factor receptor (EGFR) amplification with
sensitivity to EGFR inhibitor gefitinib in head and neck squamous
cell carcinoma cells. Clin Cancer Res. 12:4103–4111. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Herbst RS: Review of epidermal growth
factor receptor biology. Int J Radiat Oncol Biol Phys. 59(Suppl 2):
S21–S26. 2004. View Article : Google Scholar
|
|
31
|
Rocha-Lima CM, Soares HP, Raez LE and
Singal R: EGFR targeting of solid tumors. Cancer Control.
14:295–304. 2007.PubMed/NCBI
|
|
32
|
Grandis Rubin J, Melhem MF, Gooding WE,
Day R, Holst VA, Wagener MM, Drenning SD and Tweardy DJ: Levels of
TGF-alpha and EGFR protein in head and neck squamous cell carcinoma
and patient survival. J Natl Cancer Inst. 90:824–832. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Bei R, Budillon A, Masuelli L, Cereda V,
Vitolo D, Di Gennaro E, Ripavecchia V, Palumbo C, Ionna F, Losito
S, et al: Frequent overexpression of multiple ErbB receptors by
head and neck squamous cell carcinoma contrasts with rare antibody
immunity in patients. J Pathol. 204:317–325. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Tanaka K, Iyama K, Kitaoka M, Ninomiya Y,
Oohashi T, Sado Y and Ono T: Differential expression of·alpha
1(IV), alpha 2(IV), alpha 5(IV) and·alpha 6(IV) collagen chains in
the basement membrane of basal cell carcinoma. Histochem J.
29:563–570. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Bains RK, Kovacevic M, Plaster CA,
Tarekegn A, Bekele E, Bradman NN and Thomas MG: Molecular diversity
and population structure at the cytochrome P450 3A5 gene in Africa.
BMC Genet. 14:342013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Olivieri EH, da Silva SD, Mendonça FF,
Urata YN, Vidal DO, Faria MA, Nishimoto IN, Rainho CA, Kowalski LP
and Rogatto SR: CYP1A2*1C, CYP2E1*5B, and GSTM1 polymorphisms are
predictors of risk and poor outcome in head and neck squamous cell
carcinoma patients. Oral Oncol. 45:e73–e79. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Curran S and Murray GI: Matrix
metalloproteinases: Molecular aspects of their roles in tumour
invasion and metastasis. Eur J Cancer. 36:1621–1630. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Riedel F, Götte K, Schwalb J and Hörmann
K: Serum levels of matrix metalloproteinase-2 and −9 in patients
with head and neck squamous cell carcinoma. Anticancer Res.
20:3045–3049. 2000.PubMed/NCBI
|
|
39
|
Sinpitaksakul SN, Pimkhaokham A,
Sanchavanakit N and Pavasant P: TGF-β1 induced MMP-9 expression in
HNSCC cell lines via Smad/MLCK pathway. Biochem Biophys Res Commun.
371:713–718. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Aggarwal S, Takada Y, Singh S, Myers JN
and Aggarwal BB: Inhibition of growth and survival of human head
and neck squamous cell carcinoma cells by curcumin via modulation
of nuclear factor-kappaB signaling. Int J Cancer. 111:679–692.
2004. View Article : Google Scholar : PubMed/NCBI
|