Introduction
R-spondin1 (RSPO1) is a secreted protein that has
Wnt signaling-enhancing effects and is essential in gender
determination (1–3). Previous studies have demonstrated that
RSPO1 exerts proliferative effects on intestinal stem cells, and is
expected to have therapeutic applications by enhancing the host
tolerance to aggressive chemoradiotherapy and ameliorating systemic
graft-versus-host disease following allogeneic bone marrow
transplantation (4–6).
Wnt/β-catenin signaling is part of the canonical Wnt
signaling pathway and plays essential roles in the development and
maintenance of adult tissues. However, its aberrant activation is
involved in various types of human cancer (7–14). This
signaling pathway regulates gene expression by controlling the
stability of β-catenin, which is phosphorylated upon forming a
complex with adenomatous polyposis coli, Axin, casein kinase 1
(CK1) and glycogen synthase kinase 3 (GSK3) (15). The phosphorylation of β-catenin leads
to its ubiquitylation and proteasomal degradation. Binding of Wnt
to Frizzled and its coreceptor, low-density lipoprotein
receptor-related protein 6 (LRP6), along with binding of
Dishevelled, results in LRP6 phosphorylation, which in turn
activates the binding of Axin, GSK3 and CK1 to Frizzled and LRP6,
and inhibits the degradation of β-catenin (15). Stabilized β-catenin translocates to
the nucleus, where it forms a complex with the transcription factor
T-cell factor/lymphoid enhancer factor (TCF/LEF) and induces the
expression of Wnt target genes, including c-Myc, cyclin D1 and
matrix metalloproteinases (15).
Accordingly, aberrant activation of Wnt signaling is a cause of
cancer (15).
Previous experiments have demonstrated that RSPO1
enhances the activity of the Wnt/β-catenin signaling pathway
(2). Secreted RSPO1 binds
leucine-rich repeat containing G protein-coupled receptor (LGR)4 or
LGR5, in addition to the cell-surface transmembrane E3 ubiquitin
ligase zinc and ring finger 3 (ZNRF3) or its homolog, ring finger
protein 43 (RNF43) (16,17). ZNRF3 and RNF43 act as negative
regulators of Wnt/β-catenin signaling by decreasing the membrane
levels of Frizzled and LRP6 (17,18). RSPO1
induces membrane clearance of ZNRF3/RNF43 in an LGR4/5-dependent
manner and induces LRP6 phosphorylation, thus potentiating
Wnt/β-catenin signaling (16,17,19).
Glycosylation is a common post-translational
modification that is important for protein stability, folding,
secretion and a wide variety of protein functions (20–24). There
are four main types of glycosylation: N-, O- and
C-linked glycosylation, and glycosylphosphatidylinositol
anchor (25). RSPO1 contains a
consensus sequence for N-glycosylation (1), but it remains unknown whether RSPO1 is
N-glycosylated. In N-glycosylation, an
oligosaccharide chain is covalently linked to an Asn residue in the
consensus sequence Asn-Xaa-Ser/Thr (where Xaa is any amino acid
with the exception of Pro) of secreted or membrane-bound proteins
(26,27). This glycosylation is caused by a
continuous enzymatic reaction inside the lumen of the endoplasmic
reticulum (ER) and Golgi apparatus (28,29). When
a nascent glycoprotein enters the ER, a preformed N-linked
sugar chain is attached to a particular Asn in the protein
(28,29). This attached sugar chain is then
processed via the ER and the Golgi apparatus, prior to the
secretion of the N-glycosylated protein outside the cell
(28,29). The present study demonstrated that
RSPO1 is N-glycosylated at Asn137, and suggested that
N-glycosylation of RSPO1 has a negative effect on its
secretion levels and Wnt/β-catenin signaling-enhancing effects.
Materials and methods
Cell culture
The human fibrosarcoma cell line HT1080 was obtained
from the Japanese Collection of Research Bioresources Cell Bank
(Osaka, Japan) and the human embryonic kidney (HEK) cell line
HEK293T was obtained from RIKEN BioResource Center (Tsukuba,
Japan). HT1080 and HEK293T cells were cultured in Dulbecco's
modified Eagle's medium (DMEM; catalogue no. 05919; Nissui
Pharmaceutical Co., Ltd., Tokyo, Japan) supplemented with 5% (v/v)
fetal bovine serum (Bovogen Biologicals Pty Ltd., Melbourne,
Australia), 100 mg/l kanamycin (Sigma-Aldrich, St. Louis, MO, USA),
100 U/ml penicillin G (Sigma-Aldrich), 600 mg/l L-glutamine
(Sigma-Aldrich) and 2.25 g/l NaHCO3 (Wako Pure Chemical
Industries, Ltd., Osaka, Japan). Cells were incubated at 37°C in a
humidified incubator with 5% CO2.
Plasmids construction
Human complementary (c)DNA coding for RSPO1 was
amplified by polymerase chain reaction (PCR) from a cDNA library
derived from human prostate cancer PC3 cells (which was kindly
donated by Dr Nobuyuki Tanaka, Keio University School of Medicine,
Tokyo, Japan), using KOD FX Neo (Toyobo Co., Ltd., Osaka, Japan),
according to the manufacturer's protocol. The sequences of the
primers used (which were synthesized by Thermo Fisher Scientific,
Inc., Waltham, MA, USA) were as follows: Forward,
5′-TTTTCTCGAGATGCGGCTTGGGCTGTGTG-3′ and reverse,
5′-TTTTGCGGCCGCCTAGGCAGGCCCTGCAG-3′. The reaction was conducted in
a C1000™ thermal cycler (Bio-Rad Laboratories, Inc., Hercules, CA,
USA), and the cycling conditions were as follows: 94°C for 2 min,
followed by 35 cycles at 94°C for 15 sec, 63°C for 30 sec and 68°C
for 1 min. To introduce C-terminal Myc-His6 tags,
polymerase chain reaction (PCR) was performed with primers
possessing Myc and His6 codons. The sequences of the
tags were as follows: Myc, 5′-GAACAAAAACTCATCTCAGAAGAGGATCTG-3′ and
His6, 5′-CATCATCACCATCACCAT-3′. Subsequently, the
RSPO1-Myc-His6 cDNA was subcloned into the pCI-neo
vector (Promega Corporation, Madison, WI, USA). To create the
mutant N137Q RSPO1, the Asn137 residue in wild-type (wt) RSPO1 was
substituted with a Gln residue by inverse PCR in a C1000™ thermal
cycler, using wt RSPO1 plasmids as a template. Inverse PCR was
performed using KOD FX Neo according to the manufacturer's
protocol. The sequences of the primers (Thermo Fisher Scientific,
Inc., Waltham, MA, USA) used for the mutagenesis were as follows:
Forward, 5′-GCTCCTCAGCTGCCCAGGGCACCATGGAGT-3′ and reverse,
5′-ACTCCATGGTGCCCTGGGCAGCTGAGGAGC-3′. The cycling conditions used
for inverse PCR were as follows: 94°C for 2 min, followed by 20
cycles at 94°C for 15 sec, 60°C for 30 sec and 68°C for 3.5
min.
Establishment of RSPO1-overexpressing
cell line
Stable cell lines expressing wt or mutant
RSPO1-Myc-His6 were established by transfecting the
pCI-neo vectors expressing wt or N137Q mutant
RSPO1-Myc-His6 into HT1080 cells and using 400 µg/ml
G418 (Roche Applied Science, Penzberg, Germany) for the selection
of RSPO1-Myc-His6-clones. Those clones that expressed
high levels of Myc-His6-tagged wt RSPO1 and N137Q RSPO1
were named HT1080-RSPO1-MH and HT1080-RSPO1/N137Q-MH, respectively.
Cells transfected with pCI-neo were called HT1080-neo and served as
control.
Western blotting
Western blotting was performed using a slightly
modified version of a previously described protocol (30–32). Cells
were lysed in a lysis buffer [50 mM Tris (Sigma-Aldrich)-HCl (Kanto
Chemical Co., Inc., Tokyo, Japan) (pH 7.5), 150 mM NaCl (Wako Pure
Chemical Industries, Ltd.), 0.1% (w/v) sodium dodecyl sulfate (SDS;
Wako Pure Chemical Industries, Ltd.), 1% (v/v) Triton X-100 (Wako
Pure Chemical Industries, Ltd.), 1% (w/v) sodium deoxycholate (Wako
Pure Chemical Industries, Ltd.) and 1 mM phenylmethylsulfonyl
fluoride (PMSF; Sigma-Aldrich)] and homogenized with sonication (20
kHz, 50 W, 10 sec, twice) in an ultrasonic homegenizer (UH-50; SMT,
Fuji America Corporation, Vernon Hills, IL, USA). The cell lysates
were centrifuged at 18,000 × g for 10 min in an MX-307 centrifuge
(Tomy Digital Biology Co., Ltd., Tokyo, Japan), and the total
amount of protein in each lysate was measured with the Protein
Assay Dye Reagent Concentrate (Bio-Rad Laboratories, Inc.,
Hercules, CA, USA). Sample buffer 6X [350 mM Tris-HCl (pH 6.8), 30%
(w/v) glycerol (Kanto Chemical Co., Inc.), 0.012% (w/v) bromophenol
blue (Kanto Chemical Co., Inc.), 6% (w/v) SDS and 30% (v/v)
2-mercaptoethanol (2-ME; Kanto Chemical Co., Inc.)] was added to
each cell lysate, which was subsequently boiled at 98°C for 3 min
and electrophoresed on 12.5% SDS-polyacrylamide gels using a mini
gel slab electrophoresis tank (catalogue no. NA-1010; Nihon Eido
Co., Ltd., Tokyo, Japan) and a power supply (catalogue no. NC-1017;
Nihon Eido Co., Ltd.) with current and time set at 23 mA and 1.5 h,
respectively. Proteins were next transferred to polyvinylidene
fluoride membranes (GE Healthcare Life Sciences, Chalfont, UK)
using Trans-Blot SD semi-dry electrophoretic transfer cell
(catalogue no. 1703940JA, Bio-Rad Laboratories, Inc.) and a power
supply with voltage and time set at 12 V and 1 h, respectively.
Membranes were blocked with Tris-buffered saline-Tween 20 [TBST; 20
mM Tris-HCl (pH 7.6), 137 mM NaCl and 0.1% (v/v) Tween 20 (Kanto
Chemical Co., Inc.)] containing 5% Difco™ skim milk (catalogue no.
232100; BD Biosciences, Franklin Lakes, NJ, USA) for 30 min at room
temperature, and immunoblotted with rabbit polyclonal anti-c-Myc
(dilution, 1:5,000 in TBST containing 5% Difco™ skim milk;
catalogue no. C3956, Sigma-Aldrich, St. Louis, MO, USA) or mouse
monoclonal anti-α-tubulin (dilution, 1:8,000 in TBST; catalogue no.
T5168, Sigma-Aldrich) antibodies for 1 h at room temperature.
Subsequently, membranes were incubated with TBST containing 5%
Difco™ skim milk with secondary horseradish peroxidase
(HRP)-conjugated sheep polyclonal anti-mouse immunoglobulin (Ig)G
(dilution, 1:5,000; catalogue no. NA931V; GE Healthcare Life
Sciences) or donkey polyclonal anti-rabbit IgG (dilution, 1:5,000;
catalogue no. NA934V; GE Healthcare Life Sciences) antibodies for 1
h at room temperature. The membranes were washed six times with
TBST for 5 min. Detection was performed with an enhanced
chemiluminescence reagent (Immobilon Western Chemiluminescent HRP
Substrate; EMD Millipore, Billerica, MA, USA) and ImageQuant LAS
4000 mini (GE Healthcare Life Sciences). Quantification of the
protein bands was performed with ImageQuant™ TL version 8.1
software (GE Healthcare Life Sciences).
Peptide-N-glycosidase F (PNGase F)
treatment of RSPO1
PNGase F treatment was performed as previously
described, with a slight modification (22). Cells were lysed with sonication in 50
mM phosphate buffer (pH 7.5), which contained
NaH2PO4 (Kanto Chemical Co., Inc.),
Na2HPO4 (Kanto Chemical Co., Inc.), 0.1%
(w/v) SDS, 1% (v/v) Triton X-100, 1 mM PMSF and 50 mM 2-ME, and
boiled for 5 min to inactivate endogenous enzymes. Next, 0.75%
(v/v) Triton X-100 was added to the samples, which were
subsequently incubated with 0.5 U PNGase F (Roche Applied Science)
at 37°C for 3 h. The samples were next electrophoresed and analyzed
by western blotting as described above.
Semiquantitative reverse transcription
(RT)-PCR
Semiquantitative RT-PCR was performed using a
slightly modified version of a previously described protocol
(22,24,33). Total
RNA was extracted from cultured cells using TRIzol (Invitrogen;
Thermo Fisher Scientific, Inc.), according to the manufacturer's
protocol, and solutions containing 2 µg total RNA were subjected to
RT reaction using the High Capacity cDNA Reverse Transcription kit
(Applied Biosystems; Thermo Fisher Scientific, Inc.), according to
the manufacturer's protocol. The cDNAs obtained were then used for
PCR amplification with EmeraldAmp PCR Master Mix (Takara Bio, Inc.,
Otsu, Japan) in a C1000™ thermal cycler. The number of PCR cycles
for each product was determined upon confirming the efficacy of the
amplification and having defined the linear exponential phase of
the amplification. The sequences of the primers (Thermo Fisher
Scientific, Inc.), number of cycles and annealing temperatures used
for semiquantitative RT-PCR were as follows: Transfected exogenous
RSPO1, forward, 5′-CTCTGCTCTGAAGTCAACGG-3′ and reverse,
5′-GTGATGGTGATGATGCAGATCCTCTTCTGAGATGAG-3′, 25 cycles, 63°C; and
β-actin, forward, 5′-CTTCTACAATGAGCTGCGTG-3′ and reverse,
5′-TCATGAGGTAGTCAGTCAGG-3′, 20 cycles, 58°C. The PCR products were
electrophoresed on agarose gels, which were prepared in 1X
Tris/borate/ethylenediaminetetraacetic acid (EDTA) buffer [50 mM
Tris, 48.5 mM boric acid (Wako Pure Chemical Industries, Ltd.) and
2 mM EDTA (Kanto Chemical Co., Inc., Tokyo, Japan)], supplemented
with 0.5 µg/ml ethidium bromide (Sigma-Aldrich), and visualized
with an ultraviolet illuminator (Desktop Gel Imager Scope21; Optima
Inc., Tokyo, Japan).
Detection of secreted RSPO1
Cells were washed twice with phosphate-buffered
saline [PBS; containing 137 mM NaCl, 2.7 mM KCl (Kanto Chemical
Co., Inc.), 10 mM Na2HPO4 and 1.8 mM
KH2PO4] and cultured in serum-free DMEM with
50 µg/ml heparin sodium salt (catalogue no. H3149; Sigma-Aldrich)
for 24 h. Conditioned medium (CM) was then collected, and
Ni-nitrilotriacetic acid (NTA) agarose (Qiagen GmbH, Hilden,
Germany) was added to the CM, and the mixture was incubated for 2 h
at 4°C. Next, the Ni-NTA agarose was washed with washing buffer A
[900 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8
mM KH2PO4 and 20 mM imidazole (USB
Corporation, Cleveland, OH, USA)], and Ni-NTA-bound RSPO1 was
subsequently eluted with an elution buffer (900 mM NaCl, 2.7 mM
KCl, 10 mM Na2HPO4, 1.8 mM
KH2PO4 and 500 mM imidazole, pH 7.4). Upon
concentration with Ni-NTA agarose, the total amount of protein in
the CM was estimated from the total amount of protein in the cell
lysates. Samples were next electrophoresed and analyzed by western
blotting as mentioned above.
Immunofluorescence
Cells were grown on coverslips at 37°C for 24 h. To
observe the localization of the Golgi apparatus, the cells were
washed twice with PBS containing 50 µg/ml heparin sodium salt,
fixed with 4% paraformaldehyde (Wako Pure Chemical Industries,
Ltd.) for 10 min and permeabilized with 0.1% Triton X-100 for 5
min. Upon blocking with PBS containing 2% bovine serum albumin
(BSA; catalogue no. 12660; EMD Millipore), cells were incubated
with rabbit polyclonal anti-Golgi reassembly-stacking protein of 65
kDa (GRASP65) antibody (dilution, 1:100; catalogue no. sc-30093;
Santa Cruz Biotechnology, Inc., Dallas, TX, USA) and mouse
monoclonal anti-c-Myc antibody (dilution, 1:100; catalogue no.
sc-40; Santa Cruz Biotechnology, Inc.) for 1 h at room temperature.
Subsequently, cells were incubated for 1 h at room temperature with
Alexa Fluor® 568-conjugated anti-rabbit IgG (catalogue
no. A11036; Invitrogen; Thermo Fisher Scientific, Inc.) and Alexa
Fluor® 488-conjugated anti-mouse IgG (catalogue no.
A11029; Invitrogen; Thermo Fisher Scientific, Inc.) secondary
antibodies diluted 1:500 in PBS containing 2% BSA. Following two
washes with PBS, the cells were incubated with 2 µg/ml Hoechst
33258 (Polysciences, Inc., Warrington, PA, USA) for 10 min at room
temperature to stain the nuclei. The cells were then washed with
PBS and visualized under a fluorescence microscope
(EVOS® FL Cell Imaging system; Life Technologies; Thermo
Fisher Scientific, Inc.).
Purification of recombinant RSPO1 and
luciferase activity assay
Cells were washed twice with PBS and cultured in
serum-free DMEM for 24 h with 1% (v/v) Heparin Sepharose 6 Fast
Flow (GE Healthcare Life Sciences). Following 24 h, Heparin
Sepharose 6 beads were collected, washed twice with PBS and eluted
with washing buffer A. The eluted solutions were concentrated with
Ni-NTA agarose as aforementioned described, and buffer-exchanged
into PBS using Vivaspin® 500 centrifugal filter units
(Sartorius AG, Göttingen, Germany), according to the manufacture's
protocol. Purified RSPO1 was electrophoresed and detected by
western blotting as aforementioned described, and diluted RSPO1
samples were used for luciferase activity assay.
For the luciferase activity assay, HEK293T cells
were plated into 24-well plates, and 24 h later, cells were
transiently transfected with 400 ng canonical Wnt signaling
reporter Super 8× TOPFlash plasmid (firefly luciferase; catalogue
no. 12456; Addgene, Inc., Cambridge, MA, USA) or mutant reporter
Super 8× FOPFlash plasmid (firefly luciferase; catalogue no. 12457;
Addgene, Inc.) (34) and 20 ng
phRL-TK vector (Renilla luciferase; Promega Corporation) in
the presence of 10% (v/v) Wnt3A-CM produced from L Wnt3A cells
(catalogue no. CRL-2647; American Type Culture Collection,
Manassas, VA, USA), as previously described (10). Subsequently, cells were treated with
equal amounts of wt or mutant RSPO1 protein or vehicle control
(PBS). Following 24 h, cells were lysed, and firefly and
Renilla luciferase activities were respectively measured by
Luciferase Assay System (catalogue no. E1500; Promega Corporation)
and Renilla Luciferase Assay System (catalogue no. E2810;
Promega Corporation), according to the manufacturer's protocol,
using Infinite® 200 PRO microplate reader (Tecan Group
Ltd., Männedorf, Switzerland). TOPFlash and FOPFlash activities
were normalized to Renilla luciferase activity.
Statistical analysis
Statistical analyses were performed using two-tailed
Student's t-test. The results were presented as the mean ± standard
deviation. P<0.05 was considered to indicate a statistically
significant difference.
Results
RSPO1 is N-glycosylated
As human RSPO1 has one putative N-glycosylation site
at Asn137 in its amino acid sequence (Fig. 1A), the present study tested whether
RSPO1 was N-glycosylated or not. To examine the presence of
N-glycosylation in RSPO1, RSPO1-overexpressing HT1080 cells were
established (Fig. 1B). Treatment of
HT1080-RSPO1-MH cells with tunicamycin (TM; catalogue no. T7765;
Sigma-Aldrich), an inhibitor of N-glycosylation, increased the
electrophoretic mobility of RSPO1-MH, suggesting that RSPO1 was
N-glycosylated (Fig. 1B).
Next, it was assessed whether the increment of RSPO1
electrophoretic mobility by TM was the result of inhibition of
N-glycosylation. An in vitro deglycosylation assay with
PNGase F and RSPO1-MH cell lysates also resulted in a significant
reduction in the molecular weight of RSPO1-MH (Fig. 1C), which was identical to that of wt
RSPO1-MH treated with TM (Fig. 1C).
These results suggested that RSPO1 is N-glycosylated.
RSPO1 is N-glycosylated at Asn137
The present authors intended to identify the
N-glycosylation site(s) within RSPO1. To confirm that RSPO1 is
N-glycosylated at the putative N-glycosylation site Asn137
(Fig. 1A), an
N-glycosylation-defective RSPO1 mutant-expressing HT1080 cell line
was established (HT1080-RSPO1/N137Q-MH). Equal amounts of
transfected exogenous RSPO1-MH messenger (m)RNA were confirmed to
be present in the two stable cell lines by semiquantitative RT-PCR
(Fig. 2A). As expected, the N137Q
mutant RSPO1 had an increased electrophoretic mobility, compared
with wt RSPO1 (Fig. 2B), and was
identical to that of wt RSPO1 treated with 10 µg/ml TM (Fig. 2C). No effects in the molecular weight
of the protein were observed following TM treatment in cells
overexpressing N137Q mutant RSPO1 (Fig.
2C). These results demonstrated that RSPO1 is N-glycosylated
only at Asn137.
Effects of N-glycosylation of RSPO1 on
its secretion
The present study attempted to clarify the role of
N-glycosylation in the functions of RSPO1. Since the protein
expression levels of N137Q mutant RSPO1 in the cells were higher
than those of wt RSPO1 (Fig. 2B), the
effect of N-glycosylation on the secretion of RSPO1 was examined by
comparative experiments with wt and N137Q mutant
RSPO1-overexpressing cells. Surprisingly, the levels of secreted
RSPO1 were increased in N137Q mutant RSPO1-overexpressing cells,
compared with wt RSPO1-overexpressing cells (Fig. 3A and B). This result suggested that
N-glycosylation of RSPO1 negatively regulates its secretion. Next,
the intracellular localization of wt and mutant RSPO1 was
evaluated. Co-immunostaining demonstrated that both wt and N137Q
mutant RSPO1 were colocalized with the Golgi apparatus marker
GRASP65 (Fig. 3C), suggesting that
there are no differences in the intracellular localization of wt
and N137Q mutant RSPO1.
 | Figure 3.Effect of N-glycosylation on the
secretion and trafficking of RSPO1. (A) Effect of N-glycosylation
on the secretion of RSPO1. HT1080-neo, HT1080-RSPO1-MH and
HT1080-RSPO1/N137Q-MH cells were cultured in serum-free DMEM for 24
h in the presence of 50 µg/ml heparin, and CM and cell lysates were
collected. Samples from CM were concentrated with Ni-NTA agarose,
and alongside aliquots of the cell lysates, were electrophoresed
and immunoblotted with anti-c-Myc or anti-α-tubulin antibodies. (B)
Effect of N-glycosylation on the kinetics of RSPO1 secretion. Each
cell line was cultured in serum-free DMEM with 50 µg/ml heparin for
the indicated time points, and CM were collected. Following
concentration with Ni-NTA agarose, the samples were electrophoresed
and immunoblotted with anti-c-Myc antibody. Protein bands were
quantified using ImageQuant™ TL version 8.1 software to generate
the graph. Secreted levels of RSPO1 are the levels of wt RSPO1
protein quantified by the software at 24 h, which is defined as 1.
(C) Effect of N-glycosylation on the intracellular trafficking of
RSPO1. HT1080-neo, HT1080-RSPO1-MH and HT1080-RSPO1/N137Q-MH cells
were fixed prior to be stained with Hoechst 33258 (blue),
anti-c-Myc antibody (green) and anti-Golgi reassembly-stacking
protein of 65 kDa antibody (red), and observed by fluorescence
microscopy. Areas of overlapping stains indicating co-localization
were represented in yellow when the images were superimposed. Bar,
10 µm. Magnification, ×400. RSPO1, R-spondin1; neo, pCI-neo vector;
wt, wild-type; DMEM, Dulbecco's modified Eagle's medium; NTA,
nitrilotriacetic acid; CM, conditioned media. |
N-glycosylation of RSPO1 influences
Wnt/β-catenin signaling
Since previous studies have demonstrated that RSPO1
enhances Wnt/β-catenin signaling activity synergistically with Wnt
(2), the effect of N-glycosylation on
the Wnt signaling-enhancing activity of RSPO1 was examined in the
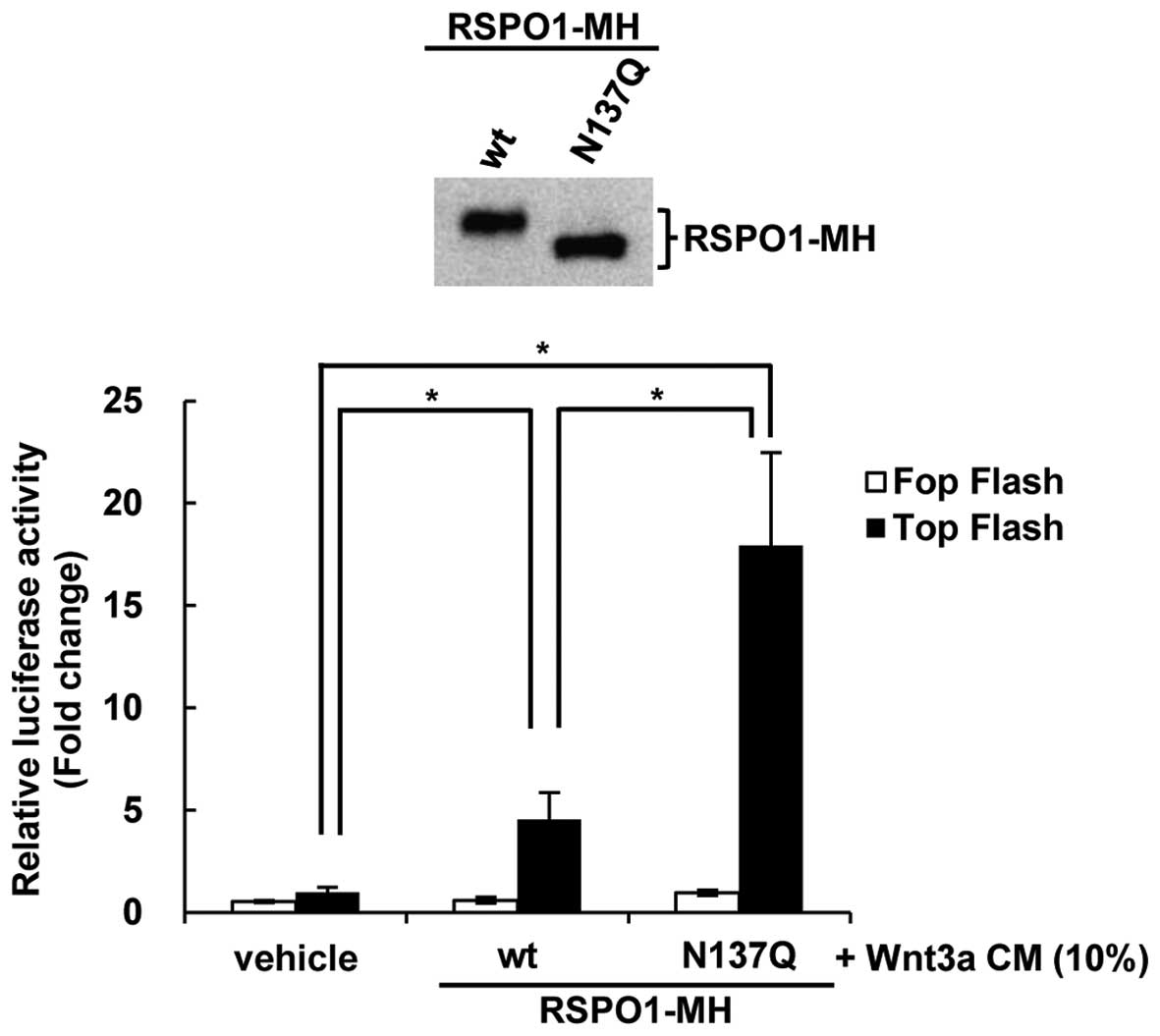
present study. Since the secreted levels of RSPO1 differed between
wt and N137Q mutant RSPO1-overexpressing cells (Fig. 3A), these recombinant RSPO1 proteins
were purified from the corresponding CM of each cell culture, and
equal amounts of RSPO1 were prepared to treat HEK293T cells for
Wnt/β-catenin signaling stimulation (Fig.
4). To determine the effects of N-glycosylation on the Wnt
signaling-enhancing activity of RSPO1, the TCF/LEF reporter plasmid
TOPFlash was used (34). As
previously reported, wt RSPO1 enhanced TOPFlash activity
synergistically with Wnt3A (10), and
N137Q mutant RSPO1 significantly increased TOPFlash activity,
compared with equivalent amounts of wt RSPO1 (P<0.05; Fig. 4). These results suggested that
N-glycosylation of RSPO1 reduces its Wnt/β-catenin
signaling-enhancing effect.
Discussion
RSPO1 is a secreted protein that exhibits Wnt
signaling-enhancing effects (1,2). However,
the role of N-glyscosylation in the function of RSPO1
remains obscure. Therefore, the present study investigated whether
RSPO1 is N-glycosylated or not, and demonstrated that RSPO1
is N-glycosylated only at Asn137, and N-glycosylation
of RSPO1 has a negative influence on its secretion and
Wnt/β-catenin signaling-enhancing effects.
Recombinant RSPO1-MH from HT1080-RSPO1-MH cell
lysates was detected as a double band of ~36.5 kDa (Fig. 1B). In contrast, the purified RSPO1-MH
from CM was detected as a single band (Fig. 3A). Therefore, it was hypothesized that
the difference in the molecular weight between RSPO1-MH in the cell
lysate and that in the CM was due to N-glycosylation, since
only N-glycosylated RSPO1 was expected to be secreted.
Contrarily to the above hypothesis, the electrophoretic mobility of
each of the bands corresponding to RSPO1-MH from cell lysates was
increased following treatment with PNGase F and TM. This result
suggested that all intracellular RSPO1 is N-glycosylated,
and that the double band of wt RSPO1 present in the cells is not
due to N-glycosylation. Therefore, further studies are
required to clarify the mechanism and roles for each of these
bands.
The present study also demonstrated that the
secreted levels of RSPO1 were increased in
non-N-glycosylated (N137Q mutant) RSPO1, compared with wt
RSPO1. Previous experiments suggested that RSPO1 binds to LGR4/5
with high affinity in the extracellular region of the cell, and is
then co-internalized with these receptors inside the cell (16). Thus, the alteration in the secreted
levels of RSPO1 due to N-glycosylation that was observed in
the present study may be explained by changes in the kinetics of
secretion, internalization and/or stability of the protein in the
extracellular region. However, the amount of RSPO1 present in the
CM was not influenced by the internalization and stability of RSPO1
in the extracellular region (data not shown). Therefore, it was
speculated that the increased secreted levels of N137Q mutant RSPO1
observed in the present study may be mainly due to an increase in
the kinetics of secretion. Indeed, the intracellular localization
of N137Q mutant RSPO1, which was present in the Golgi apparatus and
ER, was similar to that of wt RSPO1. These results suggested that
N-glycosylation of RSPO1 may regulate the transportation of
RSPO1 from the Golgi apparatus to the extracellular region, rather
than its transportation from the ER to the Golgi apparatus.
Therefore, these results suggested that the kinetics of RSPO1
secretion may be regulated by N-glycosylation. Further
studies are required for understanding the regulatory mechanisms of
RSPO1 secretion by N-glycosylation.
Furthermore, although the N137Q mutant RSPO1
increased the levels of secretion of RSPO1, the amount of
intracellular RSPO1 also increased in cells expressing this mutant.
Based on these results and the fact that the mRNA levels of
exogenous wt and N137Q mutant RSPO1 were equal, it may be
hypothesized that N-glycosylation of RSPO1 regulates the
kinetics of its secretion by influencing its intracellular
stability. Several proteins have been reported to be stabilized by
N-glycosylation (35–37), but there are limited studies reporting
whether N-glycosylation causes protein destabilization
(24). Thus, the present findings may
extend the knowledge about the role of N-glycosylation in
protein recognition and secretion.
The present study revealed that
non-N-glycosylated RSPO1 increased the Wnt/β-catenin
signaling activity significantly more than wt RSPO1 did. RSPO1
binds to heparin with high affinity, and it has been previously
suggested that secreted RSPO1 is associated with heparan sulfate
proteoglycans of the plasma membrane and extracellular matrix
(2). Therefore, the present authors
hypothesized that N-glycosylation of RSPO1 influences the
association between RSPO1 and heparin, thus affecting Wnt/β-catenin
signaling activity. However, in the present study,
N-glycosylation of RSPO1 had no effect on the association
between RSPO1 and heparin (data not shown). Previous studies have
proposed that the membrane proteins LGR4 and LGR5 recruit RSPO1 and
induce the interaction with RSPO1 and ZNRF3/RNF43, which are
antagonists of the Wnt/β-catenin signaling pathway (16,17,38). This
interaction leads to membrane clearance of ZNRF3/RNF43 and
consequently enhances the Wnt/β-catenin signaling activity
(16–18,38).
Therefore, N-glycosylation of RSPO1 is likely to affect the
association between RSPO1 and LGR4/5, and/or between RSPO1 and
ZNRF3/RNF43, thus regulating Wnt/β-catenin signaling activity.
It has been reported that aberrant
N-glycosylation causes multiple diseases (39). In addition, dysregulated activation of
Wnt/β-catenin signaling is involved a variety of human tumors
(11,12,14), and
overexpression of LGR4/5 has been reported in several types of
cancer (40,41). Therefore the N-glycosylation
status of RSPO1 may be a key regulator of cancer by aberrantly
activating Wnt/β-catenin signaling. In contrast, RSPO1 has also
been observed to exert proliferative effects on intestinal crypt
and stem cells, and is therefore expected to have therapeutic
applications (4–6). In this context, the
non-N-glycosylated RSPO1 mutant, which increased
Wnt/β-catenin signaling activity in the present study, may be more
effective than the above proposed therapeutic applications of
RSPO1.
In conclusion, the present study demonstrated that
RSPO1 is N-glycosylated at Asn137, and the
non-N-glycosylated N137Q mutant of RSPO1 exhibited increased
secretion levels and Wnt/β-catenin signaling-enhancing effects.
These results suggest that N-glycosylation of RSPO1 has a
negative influence on the secretion and Wnt/β-catenin
signaling-enhancing effects of RSPO1.
Acknowledgments
The authors would like to thank Professor Randall T.
Moon (University of Washington, Seattle, WA, USA) for kindly
donating Super 8× TOPFlash and Super 8× FOPFlash plasmids.
The present study was partly supported by grants
from the programs Grants-in-Aid for Scientific Research (B) [Japan
Society for the Promotion of Science (JSPS); grant no. 24310167)
and JSPS Fellows (grant no. 254256). Mr. Yuki Niwa is a JSPS
Research Fellow.
Glossary
Abbreviations
Abbreviations:
|
RSPO
|
R-spondin
|
|
CK1
|
casein kinase 1
|
|
GSK3
|
glycogen synthase kinase 3
|
|
LRP6
|
low-density lipoprotein
receptor-related protein 6
|
|
TCF/LEF
|
T-cell factor/lymphoid enhancer
factor
|
|
LGR
|
leucine-rich repeat containing G
protein-coupled receptor
|
|
ZNRF3
|
zinc and ring finger 3
|
|
RNF43
|
ring finger protein 43
|
|
ER
|
endoplasmic reticulum
|
|
DMEM
|
Dulbecco's modified Eagle's medium
|
|
PNGase F
|
peptide-N-glycosidase F
|
|
PBS
|
phosphate-buffered saline
|
|
TM
|
tunicamycin
|
References
|
1
|
Kamata T, Katsube K, Michikawa M, Yamada
M, Takada S and Mizusawa H: R-spondin, a novel gene with
thrombospondin type 1 domain, was expressed in the dorsal neural
tube and affected in Wnts mutants. Biochim Biophys Acta.
1676:51–62. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Nam JS, Turcotte TJ, Smith PF, Choi S and
Yoon JK: Mouse cristin/R-spondin family proteins are novel ligands
for the Frizzled 8 and LRP6 receptors and activate
beta-catenin-dependent gene expression. J Biol Chem.
281:13247–13257. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Parma P, Radi O, Vidal V, Chaboissier MC,
Dellambra E, Valentini S, Guerra L, Schedl A and Camerino G:
R-spondin1 is essential in sex determination, skin differentiation
and malignancy. Nat Genet. 38:1304–1309. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kim KA, Kakitani M, Zhao J, Oshima T, Tang
T, Binnerts M, Liu Y, Boyle B, Park E, Emtage P, et al: Mitogenic
influence of human R-spondin1 on the intestinal epithelium.
Science. 309:1256–1259. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Takashima S, Kadowaki M, Aoyama K, Koyama
M, Oshima T, Tomizuka K, Akashi K and Teshima T: The Wnt agonist
R-spondin1 regulates systemic graft-versus-host disease by
protecting intestinal stem cells. J Exp Med. 208:285–294. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhou WJ, Geng ZH, Spence JR and Geng JG:
Induction of intestinal stem cells by R-spondin 1 and Slit2
augments chemoradioprotection. Nature. 501:107–111. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Korinek V, Barker N, Moerer P, van
Donselaar E, Huls G, Peters PJ and Clevers H: Depletion of
epithelial stem-cell compartments in the small intestine of mice
lacking Tcf-4. Nat Genet. 19:379–383. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kuhnert F, Davis CR, Wang HT, Chu P, Lee
M, Yuan J, Nusse R and Kuo CJ: Essential requirement for Wnt
signaling in proliferation of adult small intestine and colon
revealed by adenoviral expression of Dickkopf-1. Proc Natl Acad Sci
USA. 101:266–271. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Pinto D, Gregorieff A, Begthel H and
Clevers H: Canonical Wnt signals are essential for homeostasis of
the intestinal epithelium. Genes Dev. 17:1709–1713. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Willert K, Brown JD, Danenberg E, Duncan
AW, Weissman IL, Reya T, Yates JR III and Nusse R: Wnt proteins are
lipid-modified and can act as stem cell growth factors. Nature.
423:448–452. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Giles RH, van Es JH and Clevers H: Caught
up in a Wnt storm: Wnt signaling in cancer. Biochim Biophys Acta.
1653:1–24. 2003.PubMed/NCBI
|
|
12
|
Lammi L, Arte S, Somer M, Jarvinen H,
Lahermo P, Thesleff I, Pirinen S and Nieminen P: Mutations in AXIN2
cause familial tooth agenesis and predispose to colorectal cancer.
Am J Hum Genet. 74:1043–1050. 2004. View
Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kinzler KW, Nilbert MC, Su LK, Vogelstein
B, Bryan TM, Levy DB, Smith KJ, Preisinger AC, Hedge P and
McKechnie D: Identification of FAP locus genes from chromosome
5q21. Science. 253:661–665. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Satoh S, Daigo Y, Furukawa Y, Kato T, Miwa
N, Nishiwaki T, Kawasoe T, Ishiguro H, Fujita M, Tokino T, et al:
AXIN1 mutations in hepatocellular carcinomas and growth suppression
in cancer cells by virus-mediated transfer of AXIN1. Nat Genet.
24:245–250. 2000. View
Article : Google Scholar : PubMed/NCBI
|
|
15
|
MacDonald BT, Tamai K and He X:
Wnt/β-catenin signaling: Components, mechanisms, and diseases. Dev
Cell. 17:9–26. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Carmon KS, Gong X, Lin Q, Thomas A and Liu
Q: R-spondins function as ligands of the orphan receptors LGR4 and
LGR5 to regulate Wnt/β-catenin signaling. Proc Natl Acad Sci USA.
108:11452–11457. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Hao HX, Xie Y, Zhang Y, Charlat O, Oster
E, Avello M, Lei H, Mickanin C, Liu D, Ruffner H, et al: ZNRF3
promotes Wnt receptor turnover in an R-spondin-sensitive manner.
Nature. 485:195–200. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Koo BK, Spit M, Jordens I, Low TY, Stange
DE, van de Wetering M, van Es JH, Mohammed S, Heck AJ, Maurice MM
and Clevers H: Tumour suppressor RNF43 is a stem-cell E3 ligase
that induces endocytosis of Wnt receptors. Nature. 488:665–669.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wei Q, Yokota C, Semenov MV, Doble B,
Woodgett J and He X: R-spondin1 is a high affinity ligand for LRP6
and induces LRP6 phosphorylation and beta-catenin signaling. J Biol
Chem. 282:15903–15911. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Dwek RA, Butters TD, Platt FM and Zitzmann
N: Targeting glycosylation as a therapeutic approach. Nat Rev Drug
Discov. 1:65–75. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
21
|
Simizu S, Ishida K, Wierzba MK and Osada
H: Secretion of heparanase protein is regulated by glycosylation in
human tumor cell lines. J Biol Chem. 279:2697–2703. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Niwa Y, Suzuki T, Dohmae N, Umezawa K and
Simizu S: Determination of cathepsin V activity and intracellular
trafficking by N-glycosylation. FEBS Lett. 586:3601–3607. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Goto Y, Niwa Y, Suzuki T, Dohmae N,
Umezawa K and Simizu S: C-mannosylation of human
hyaluronidase 1: Possible roles for secretion and enzymatic
activity. Int J Oncol. 45:344–350. 2014.PubMed/NCBI
|
|
24
|
Uematsu S, Goto Y, Suzuki T, Sasazawa Y,
Dohmae N and Simizu S: N-glycosylation of extracellular
matrix protein 1 (ECM1) regulates its secretion, which is unrelated
to lipoid proteinosis. FEBS Open Bio. 4:879–885. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Spiro RG: Protein glycosylation: Nature,
distribution, enzymatic formation, and disease implications of
glycopeptide bonds. Glycobiology. 12:43R–56R. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Blom N, Sicheritz-Pontén T, Gupta R,
Gammeltoft S and Brunak S: Prediction of post-translational
glycosylation and phosphorylation of proteins from the amino acid
sequence. Proteomics. 4:1633–1649. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Gavel Y and von Heijne G: Sequence
differences between glycosylated and non-glycosylated Asn-X-Thr/Ser
acceptor sites: Implications for protein engineering. Protein Eng.
3:433–442. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ruddock LW and Molinari M: N-glycan
processing in ER quality control. J Cell Sci. 119:4373–4380. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kornfeld R and Kornfeld S: Assembly of
asparagine-linked oligosaccharides. Annu Rev Biochem. 54:631–664.
1985. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Yasukagawa T, Niwa Y, Simizu S and Umezawa
K: Suppression of cellular invasion by glybenclamide through
inhibited secretion of platelet-derived growth factor in ovarian
clear cell carcinoma ES-2 cells. FEBS Lett. 586:1504–1509. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Kuroda M, Funasaki S, Saitoh T, Sasazawa
Y, Nishiyama S, Umezawa K and Simizu S: Determination of
topological structure of ARL6ip1 in cells: Identification of the
essential binding region of ARL6ip1 for conophylline. FEBS Lett.
587:3656–3660. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Ukaji T, Sasazawa Y, Umezawa K and Simizu
S: Involvement of conserved tryptophan residues for secretion of
TIMP-2. Oncol Lett. 7:631–634. 2014.PubMed/NCBI
|
|
33
|
Goto Y, Niwa Y, Suzuki T, Uematsu S,
Dohmae N and Simizu S: N-glycosylation is required for
secretion and enzymatic activity of human hyaluronidase1. FEBS Open
Bio. 4:554–559. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Veeman MT, Slusarski DC, Kaykas A, Louie
SH and Moon RT: Zebrafish prickle, a modulator of noncanonical
Wnt/Fz signaling, regulates gastrulation movements. Curr Biol.
13:680–685. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Solá RJ and Griebenow K: Effects of
glycosylation on the stability of protein pharmaceuticals. J Pharm
Sci. 98:1223–1245. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Waetzig GH, Chalaris A, Rosenstiel P,
Suthaus J, Holland C, Karl N, Vallés Uriarte L, Till A, Scheller J,
Grötzinger J, et al: N-linked glycosylation is essential for
the stability but not the signaling function of the interleukin-6
signal transducer glycoprotein 130. J Biol Chem. 285:1781–1789.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Zou S, Huang S, Kaleem I and Li C:
N-glycosylation enhances functional and structural stability of
recombinant β-glucuronidase expressed in Pichia pastoris. J
Biotechnol. 164:75–81. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
de Lau W, Peng WC, Gros P and Clevers H:
The R-spondin/Lgr5/Rnf43 module: Regulator of Wnt signal strength.
Genes Dev. 28:305–316. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Cylwik B, Naklicki M, Chrostek L and
Gruszewska E: Congenital disorders of glycosylation. Part I.
Defects of protein N-glycosylation. Acta Biochim Pol. 60:151–161.
2013.PubMed/NCBI
|
|
40
|
Nakata S, Phillips E and Goidts V:
Emerging role for leucine-rich repeat-containing G-protein-coupled
receptors LGR5 and LGR4 in cancer stem cells. Cancer Manag Res.
6:171–180. 2014.PubMed/NCBI
|
|
41
|
Chen Q, Cao HZ and Zheng PS: LGR5 promotes
the proliferation and tumor formation of cervical cancer cells
through the Wnt/β-catenin signaling pathway. Oncotarget.
5:9092–9105. 2014. View Article : Google Scholar : PubMed/NCBI
|