Introduction
Esophageal cancer is one of the most common types of
cancer worldwide with >480,000 new cases diagnosed annually
(1,2).
Globally, the disease accounts for ~400,000 cancer-associated
mortalities annually (3) with a
5-year survival rate of <20% (4).
According to the National Comprehensive Cancer Network guidelines
(5), surgery is the optimal treatment
choice, however, chemotherapy and radiotherapy may also be
administered (6). Squamous cell
carcinoma is one of the most common types of cancer and this reacts
poorly to common chemotherapy compared with other types of cancer,
such as adenocarcinoma (7). Novel
studies are required to provide evidence for effective therapy on
this disease. Glycolysis and the uptake of glucose are enhanced in
cancer cells in order to meet the increased energy requirements of
the rapidly proliferating cells, including esophageal caner cells,
which are known of elevated Wurberg Effects. A fraction of the
glucose in cancer cells is metabolized by the hexosamine
biosynthetic pathway (HBP) (8–10). The HBP
regulates enzymatic O-linked glycosylation (O-GlcNAcylation), a
post-translational carbohydrate modification of diverse nuclear and
cytosolic proteins by the addition of O-linked
β-N-acetylglucosamine (O-GlcNAc). O-GlcNAcylation of a protein
alters the protein's stability, intracellular localization and
function. O-GlcNAcylation serves important roles in an array of
normal biological processes, and its dysregulation is involved in a
variety of human diseases, including diabetes mellitus (11,12) and
various neurological disorders (13).
Several GlcNAcylated tumor-associated proteins have recently been
identified, including c-Myc (14) and
p53 (15), each one of the most
important oncogenes and tumor suppressor genes, respectively. These
findings suggest that O-GlcNAcylation serves a significant role in
oncogenesis and tumor progression (16–18). In
our previous study, OGT and a marker of O-GlcNAcylation levels,
mouse monoclonal anti-O-linked N-acetylglucosamine antibody (RL2),
were observed to be upregulated in esophageal cancer (19). However, the physiological consequences
of this upregulation remain to be determined.
In the current study, RNA interference (RNAi) was
used to knock down OGT in esophageal cancer Eca-109 cells, and the
cell proliferation and migration capabilities were subsequently
assessed. The results demonstrated that cell viability was
unaffected by RNAi knockdown of OGT in Eca-109 esophageal cancer
cells; however, the knockdown significantly reduced cell migration
and markedly decreased matrix metalloproteinase 9 (MMP9) levels in
knockdown cells.
Materials and methods
Reagents
Polyclonal rabbit anti-human OGT antibody was
purchased from ProteinTech Group, Inc. (Chicago, IL, USA; catalog
no., 11576-2-AP) and mouse monoclonal anti-human MMP9 antibody was
purchased from Santa Cruz Biotechnology, Inc. (Dallas, TX, USA;
catalog no., sc-12759). Mouse monoclonal RL2 antibody (catalog no.,
MA1-072) and the Invitrogen™ BLOCK-iT™ Pol II miR RNAi
Expression Vector Kit (catalog no., K4935-00) were
purchased from Thermo Fisher Scientific, Inc. (Waltham, MA, USA).
Transwell plates were purchased from Corning Incorporated (Corning,
NY, USA).
Cell culture and RNAi
The human esophageal cancer Eca-109 cell line was
obtained from the Type Culture Collection of the Chinese Academy of
Sciences (Shanghai, China) and cultured in RPMI-1640 medium (Thermo
Fisher Scientific, Inc.) containing 10% fetal bovine serum (FBS;
HyClone; GE Healthcare Life Sciences, Logan, UT, USA), 100 U/ml
penicillin and 100 µg/ml streptomycin (Gibco; Thermo Fisher
Scientific, Inc.). Cells were incubated at 37°C in an atmosphere of
5% CO2. Three shRNAs were designed with the assistance
of the online RNAi software BLOCK-iT™ RNAi Designer (http://rnaidesigner.lifetechnologies.com/rnaiexpress/).
The names and sequences of the three shRNA segments are listed in
Table I. Eca-109 cells were
transfected with an RNAi-OGT-EmGFP Vector according to the
BLOCK-iT™ Pol II miR RNAi Expression Vector Kit manufacturer's
instructions. Negative control cells were composed of Eca-109 cells
transfected with negative control shRNA, as described in Table I.
 | Table I.shRNA segment names and sequences. |
Table I.
shRNA segment names and sequences.
|
| Sequence |
|---|
|
|
|
|---|
| shRNA segment | Top strand | Bottom strand |
|---|
| RNAi-OGTA |
5′-TGCTGGATGTGCCAACTCAGC |
5′-CCTGGATGTGCCAACAGCTAA |
|
|
TAACCGTTTTGGCCACTGACTG |
CCGTCAGTCAGTGGCCAAAACG |
|
|
ACGGTTAGCTGTTGGCACATC-3′ |
GTTAGCTGAGTTGGCACATCC-3′ |
| RNAi-OGTB |
5′-TGCTGTAGAGTAGGCATCAGC |
5′-CCTGTAGAGTAGGCAAGCAA |
|
|
AAAGGGTTTTGGCCACTGACTG |
AGGGTCAGTCAGTGGCCAAAAC |
|
|
ACCCTTTGCTTGCCTACTCTA-3′ |
CCTTTGCTGATGCCTACTCTAC-3′ |
| RNAi-OGTC |
5′-TGCTGAATACTGCTCAGCAAC |
5′-CCTGAATACTGCTCAAACTTC |
|
|
TTCAGGTTTTGGCCACTGACTG |
AGGTCAGTCAGTGGCCAAAACC |
|
|
ACCTGAAGTTTGAGCAGTATT-3′ |
TGAAGTTGCTGAGCAGTATTC-3′ |
| Negative
control |
5′-GAAATGTACTGCGCGTGGAG |
|
|
|
ACGTTTTGGCCACTGACTGACG |
|
|
|
TCTCCACGCAGTACATTT-3′ |
|
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was extracted from the Eca-109 cells using
TRIzol reagent according to the manufacturers' instructions (Takara
Bio, Otsu, Japan), and cDNA was synthesized using GoScript™ Reverse
Transcription System, according to the manufacturer's instructions
(Promega Corporation, Madison, WI, USA; catalog no., A5001). In
total, 950 ng of total RNA from each sample was used for the
synthesis of cDNA. The target gene mRNA levels relative to the
GAPDH control were determined by qPCR in an Applied Biosystems™
7900 HT Fast Real-Time PCR System (Thermo Fisher Scientific, Inc.)
using the GoTaq® qPCR Master Mix (Promega Corporation;
catalog no., A6001). The data were analyzed using the
2−ΔΔCq method (20). The
genes of interest were OGT1, OGT2, OGT3 and MMP9. The primers were
designed using BLOCK-iT™ RNAi Designer software (Invitrogen; Thermo
Fisher, Inc.) and synthesized by BGI Shenzhen (Shenzhen, China).
The following primer sequences were used: MMP9, forward
5′-CCTGGAGACCTGAGAACCAATCT-3′ and reverse
5′-CCACCCGAGTGTAACCATAGC-3′. PCR was performed under the following
conditions: Initial denaturation at 95°C for 30 sec, followed by 40
cycles of 95°C for 5 sec, and elongation at 60°C for 30 sec. Each
of the cDNA samples from each group were assessed for gene
expression in duplicate. All the tests were repeated for 3
times.
Western blot
Total protein was extracted on ice from Eca-109
cells with cell lysis buffer (Cell Signaling Technology, Inc.,
Danvers, MA, USA). Equal amounts of protein were separated by 10%
SDS-PAGE and then transferred to a polyvinylidene difluoride
membrane (catalog no., P2813; Sigma-Aldrich, St. Louis, MO, USA).
Membranes were blocked with 2% fat-free milk in phosphate-buffered
saline (PBS) at room temperature for 1 h and then probed with
primary antibodies (dilution, 1:200) overnight at 4°C in PBS
containing 0.1% Tween 20 (PBST) and 1% fat-free milk. Following the
overnight incubation, membranes were washed four times in PBST and
then incubated with anti-rabbit horseradish peroxidase-conjugated
secondary antibody (dilution, 1:200; catalog no., 1662408; Bio-Rad
Laboratories, Inc., Hercules, CA, USA). Signals were developed
using enhanced chemiluminescence reagents (Amersham Pharmacia
Biotech, Piscataway, NJ, USA). Densitometric analysis was performed
to quantify the results using Image Pro Plus 6.0 software (Media
Cybernetics, Inc., Rockville, MD, USA).
Cell proliferation assay
Cells were seeded into 96-well plates at a density
of 5×104/ml in 100 µl of medium, and grown for 24 h.
Following transfection for 24–72 h, the CellTiter 96®
AQueous One Solution Cell Proliferation Assay System (Promega
Corporation) was added (20 µl/well) and incubated for 90 min.
Finally, the optical densities (OD) were measured at 492 nm with a
scanning microplate reader (EnSpire® Multimode Plate
Reader; Perkin Elmer, Waltham, MA, USA).
Colony formation assay
A total of 200 cells were plated in a 10-cm Petri
dish. Two weeks following transfection, the cells were washed with
PBS, fixed with cold methanol for 15 min at −20°C, and stained with
1% crystal violet (Thermo Fisher Scientific, Inc.) in 25% methanol
for 15 min. The dishes were thoroughly washed with water, and the
blue colonies were counted.
Transwell chamber assay
Transwell chamber migration assays were performed
using Nunc 24-well 8.0-µm pore Transwell plates (Thermo Fisher
Scientific, Inc.), according to the manufacturer's instructions.
Eca-109 cells were plated at a density of 5×104cells/ml
in each well with RPMI-1640 medium (Gibco; Thermo Fisher
Scientific, Inc.) free of FBS, and 500 µl culture medium containing
10% FBS was added to the bottom of the 24-well plate. Following
incubation for 24 h, non-invading cells were removed from the upper
surface of the membrane using a cotton-tipped swab. The invading
cells were subsequently fixed in methanol for 10 min and stained
with 0.1% crystal violet hydrate (Sigma-Aldrich) for 30 min. The
stained cells were counted as cells per field at 10× magnification
in 3 fields (CX31 microscope; Olympus Corporation, Tokyo,
Japan).
Statistical analysis
All data are presented as the mean ± standard error
of the mean of at least three individual experiments. Student's
t-test was used to analyze the differences between groups.
Statistical significance was determined using SPSS software version
13.0 (SPSS Inc., Chicago, IL, USA). P<0.05 was considered to
indicate statistically significant differences.
Results
Silencing of OGT by RNAi in Eca-109
cells
Three pairs of double-stranded OGT oligonucleotides
(RNAi-OGTA, RNAi-OGTB and RNAi-OGTC) were designed and cloned into
the transfection vector RNAi-OGT-EmGFP. To assess the transfection
efficiency, Eca-109 cells were transfected with RNAi-OGT-EmGFP and
EmGFP fluorescence was observed after 7-, 14- and 21-day
incubations. After 7 days of incubation, >90% of the cells
fluoresced green, and the cells remained strongly fluorescent until
at least 21 days (Fig. 1A),
indicating efficient transfection of the construct.
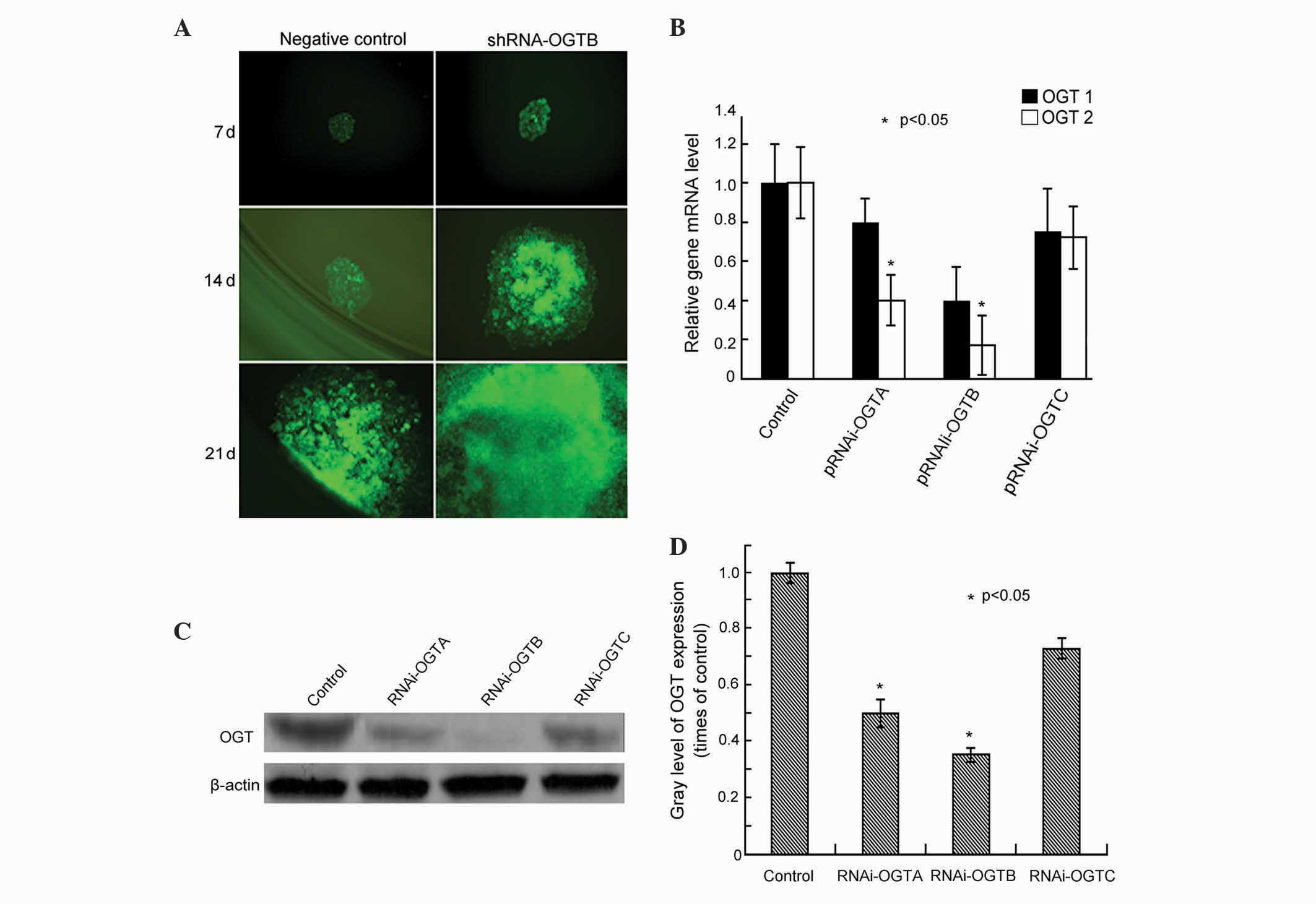 | Figure 1.Effects of transfection of RNAi-OGT in
Eca-109 esophageal cancer cells. (A) Eca-109 cells were transfected
with RNAi-OGT-EmGFP (RNAi-OGTA, RNAi-OGTB or RNAi-OGTC) for 7, 14
and 21 days, and EmGFP fluorescence was assessed using fluorescence
microscopy. Representative fluorescent images of
RNAi-OGTB-transfected cells (green) are shown. (B) Eca-109 cells
were transfected with RNAi-OGTA, RNAi-OGTB or RNAi-OGTC for 72 h,
and OGT mRNA levels of were determined by reverse
transcription-quantitative polymerase chain reaction. The RNAi
transfected cells had a significantly decreased expression of OGTA
and OGTB compared with the negative control cells. OGT1, cells
transfected with negative control shRNA; OGT2, cells transfected
with RNAi-OGTA, RNAi-OGTB or RNAi-OGTC. (C) Eca-109 cells were
transfected with RNAi-OGTA, RNAi-OGTB or RNAi-OGTC for 72 h, and
OGT protein levels were determined by western blotting with β-actin
as the loading control. (D) Gradation value ratio of OGT on western
blotting (C) demonstrated that the RNAi transfected cells had a
significantly decreased expression of OGTA and OGTAB compared with
the negative control cells. Gray levels calculated following
densitometric analysis of western blots. *P<0.05 vs. cells
infected with negative control shRNA. RNAi, RNA interference; OGT,
O-linked N-acetylglucosamine transferase. |
To assess the RNAi efficiency of the RNAi-OGT
constructs, the OGT mRNA levels were measured using RT-qPCR
following three days of transfection. The results indicated that
RNAi-OGTA, RNAi-OGTB and RNAi-OGTC decreased OGT mRNA levels
compared with negative control shRNA transfected cells.
Furthermore, the strongest OGT knockdown was induced by RNAi-OGTB
(Fig. 1B). Consistent with OGT mRNA
level alterations, western blot analysis demonstrated that OGT
protein levels were also markedly downregulated by RNAi-OGTA,
RNAi-OGTB and RNAi-OGTC. Once again, the strongest OGT knockdown
was induced by RNAi-OGTB (Fig. 1C and
D). Therefore, the subsequent functional analyses of OGT
knockdown in Eca-109 cells were performed using the RNAi-OGTB
construct.
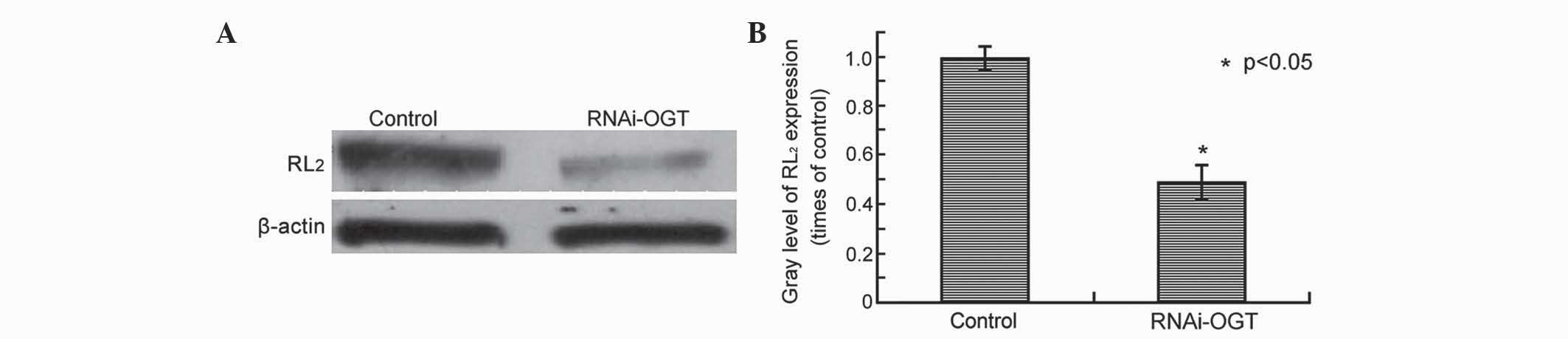
OGT knockdown by RNAi decreases
O-GlcNAcylation in Eca-109 cells
In our previous study, OGT and the O-GlcNAcylation
marker RL2 were observed to be highly expressed in esophageal
cancer (19). The present study aimed
to determine whether RNAi silencing of OGT leads to decreased
O-GlcNAcylation in esophageal cancer cells. RL2 protein levels were
assessed by western blot analysis of Eca-109 cells transfected with
RNAi-OGTB for 72 h, revealing that, compared with cells transfected
with the control shRNA, RNAi-OGTB-transfected cells exhibited
significantly decreased RL2 protein levels (P=0.002; Fig. 2).
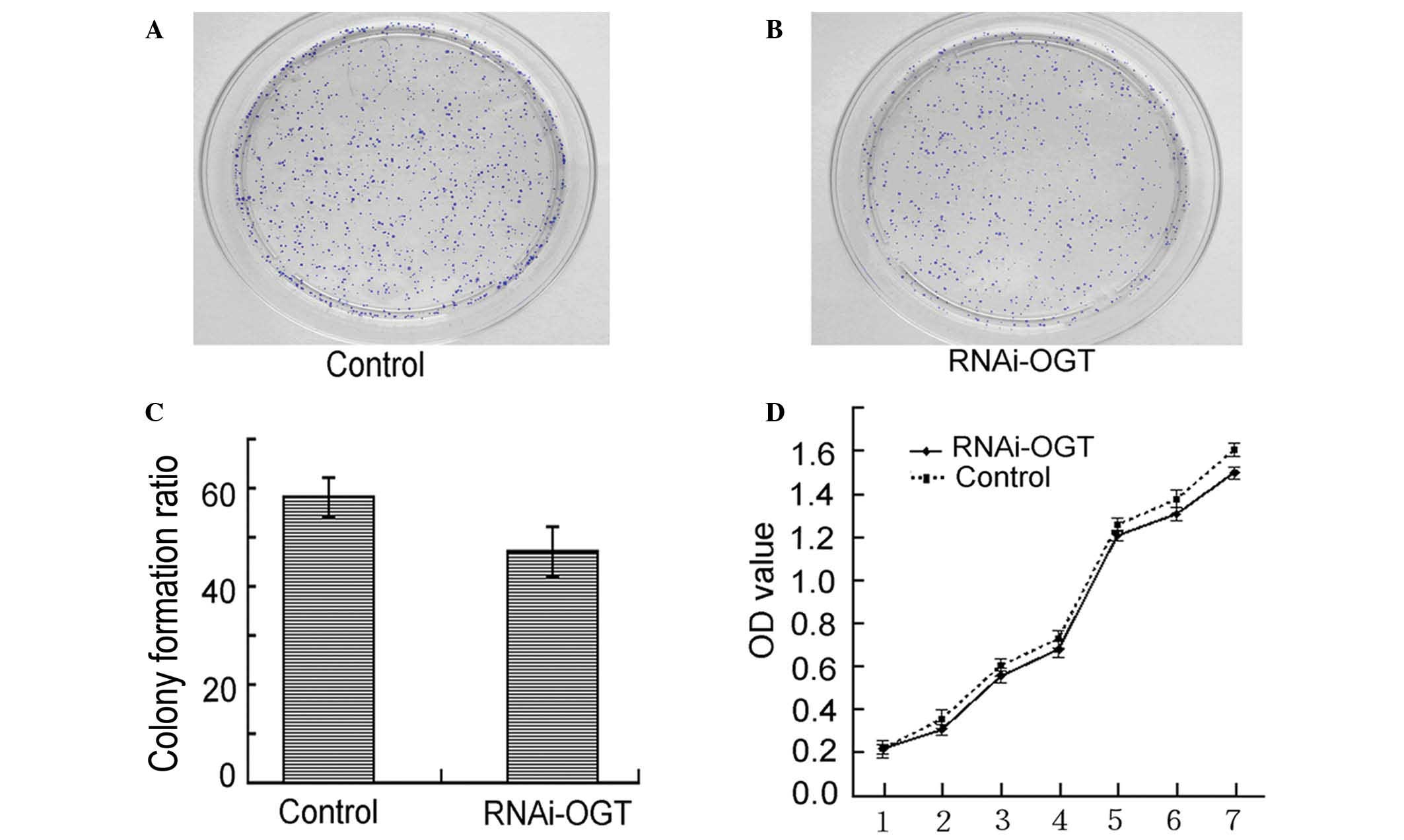
RNAi knockdown of OGT does not affect
the viability of Eca-109 cells
Colony formation assays demonstrated that RNAi-OGTB
transfection of Eca-109 cells led to decreased colony formation.
However, the observed decrease in colony formation did not reach
statistical significance when compared with the negative control
shRNA transfected cells (P=0.232; Fig.
3A–C). Similarly, cell proliferation assays revealed that
RNAi-OGTB transfection of Eca-109 cells did not significantly
inhibit cell proliferation when compared with negative control
shRNA transfected cells (P=0.728; Fig.
3D). These results indicate that the RNAi knockdown of OGT had
no effect on Eca-109 cell viability.
RNAi knockdown of OGT inhibits Eca-109
cell migration
To investigate whether RNAi knockdown of OGT affects
cell migration, Transwell assays were performed using Eca-109 cells
transfected with RNAi-OGTB for 72 h. In comparison with the control
(Fig. 4A), RNAi-OGTB transfection
significantly decreased cell migration (P<0.001; Fig. 4B and C), suggesting that OGT promotes
cell migration.
RNAi knockdown of OGT downregulates
MMP9 expression in Eca-109 cells
To investigate the underlying mechanism by which OGT
downregulation inhibits cell migration, MMP9 mRNA and protein
levels were assessed by RT-qPCR and western blot analysis,
respectively, in Eca-109 cells transfected with RNAi-OGTB for 72 h.
Compared to the control, RNAi-OGTB transfection significantly
downregulated MMP9 mRNA levels (P<0.001; Fig. 5A). Correspondingly, western blot
analysis also demonstrated the marked downregulation of MMP9 by
RNAi-OGTB transfection (P=0.003; Fig. 5B
and C).
Discussion
In this study, OGT was successfully knocked down
using RNAi in human Eca-109 esophageal cancer cells. OGT
downregulation resulted in a decrease of the O-GlcNAcylation marker
RL2. OGT knockdown did not significantly alter cell viability;
however, it did significantly reduce Eca-109 cell migration.
Moreover, MMP9 mRNA and protein expression was significantly
decreased following RNAi knockdown of OGT in Eca-109 cells. These
findings suggest that OGT may promote the migration, invasion and
metastasis of esophageal cancer cells by enhancing the stability or
expression of MMP9.
Types of protein modification include
phosphorylation, ubiquitination, glycosylation, nitrosylation and
O-GlcNAcylation. O-GlcNAcylation is a kind of post-translational
modification that targets diverse nuclear and cytosolic proteins by
the cycling of a single O-linked β-N-acetylglucosamine on the
hydroxyl groups of the serine and threonine residues of target
proteins (17,18,21).
O-GlcNAcylation is dynamically regulated by the polypeptides OGT
and β-N-acetylglucosaminidase (OGA): OGT catalyzes the addition of
O-linked β-N-acetylglucosamine from uridine diphosphate
N-acetylglucosamine onto the hydroxyl group of a serine or
threonine residue on the target protein substrate (22), whilst OGA is a neutral hexosaminidase
with a catalytic site similar to a family of 84 glycoside
hydrolases that specifically catalyze the removal of β-linked
GlcNAc on their substrates (23–25).
O-GlcNAcylation is involved in cellular signal transduction
pathways and regulates cell growth, proliferation and migration
(26). Increased OGT expression and
O-GlcNAcylation levels have been observed to promote tumorigenesis
in numerous types of tissue, including lung, colon and breast
cancers (27,28). Our group previously demonstrated that
the expression of OGT was positively related to the level of
O-GlcNAcylation, and that OGT expression and O-GlcNAcylation levels
were increased in esophageal cancer when compared to normal
esophageal tissues (19). These
results led us to hypothesize that elevated OGT expression may
promote the tumorigenesis and progression of esophageal cancer. To
investigate this hypothesis, an RNAi knockdown of OGT in the
Eca-109 human esophageal cell line was created.
The results of the present study demonstrated that
neither the RNAi knockdown of OGT expression nor the decrease of
O-GlcNAcylation significantly inhibited cellular proliferation.
This negative result may be due to the incomplete depletion of OGT
in these cells; in previous studies, complete deletion of OGT led
to cell death (29,30). High levels of O-GlcNAcylation in
breast cancer have recently been observed to promote metastasis,
particularly to lymph nodes (31).
Furthermore, in our previous report, high levels of O-GlcNAcylation
were identified to be associated with lymph node metastasis in
esophageal carcinoma (19). These
results suggest that OGT may promote cancer cell metastasis. In
accordance with this suggestion, the Transwell assay performed in
the current study demonstrated that RNAi knockdown of OGT
significantly reduced cell migration. Since matrix
metalloproteinases (MMPs), including MMP9, serve crucial roles in
cell migration (32,33), the effect of RNAi knockdown of OGT in
Eca-109 cells on MMP9 expression was investigated. This revealed
that MMP9 was significantly decreased in the RNAi-OGTB-transfected
Eca-109 cells when compared with negative control shRNA transfected
cell group, suggesting that knocking down the OGT gene in
esophageal cancer may affect cellular migratory ability by
decreasing MMP9, a phenomenon which may be predicted to be relative
to the level of O-GlcNAcylation. Supporting this hypothesis, high
OGT expression and high levels of O-GlcNAcylation have been
observed to promote the metastasis of breast cancer cells (34). Also consistent with the current
results, MMP9 has been reported to be involved in esophageal
carcinoma metastasis (35), and high
levels of O-GlcNAcylation have been associated with lymph node
metastasis in esophageal carcinoma (19). However, the molecular mechanism by
which O-GlcNAcylation affects MMP9 is largely unknown.
O-GlcNAcylation has been reported to regulate extracellular
signal-regulated kinases (ERKs) through MAPK/ERK kinases and Raf
(36), and ERK regulates MMP2 and
MMP9 (37,38). These facts imply a potential
mechanism, in which OGT knockdown results in the downregulation of
O-GlcNAcylation, which in turn results in the downregulation of
MMP9 through the ERK pathway.
In summary, the current study demonstrated that RNAi
is able to successfully downregulate OGT, and that OGT
downregulation suppresses O-GlcNAcylation in Eca-109 cells. OGT
knockdown did not affect cell viability; however, it did
significantly reduce cell migration. Supporting this result, OGT
knockdown was accompanied by a decrease in MMP9 expression. These
findings suggest that O-GlcNAcylation may promote the migration,
invasion and metastasis of esophageal cancer cells by enhancing the
stability or expression of MMP9.
Acknowledgements
The present study was funded by the Scientific and
Technological Research of Shaanxi Province of China (grant no.
2007K09-01), and was approved by the Ethics Review Committee of the
Second Affiliated Hospital of Xi'an Jiaotong University (Xi'an,
China).
References
|
1
|
Ferlay J, Shin HR, Bray F, Forman D,
Mathers C and Parkin DM: Estimates of worldwide burden of cancer in
2008: GLOBOCAN 2008. Int J Cancer. 127:2893–2917. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Pakzad R, Mohammadian-Hafshejani A,
Khosravi B, Soltani S, Pakzad I, Mohammadian M, Salehiniya H and
Momenimovahed Z: The incidence and mortality of esophageal cancer
and their relationship to development in Asia. Ann Transl Med.
4:292016.PubMed/NCBI
|
|
3
|
Parkin DM, Bray FI and Devesa SS: Cancer
burden in the year 2000. The global picture. Eur J Cancer. 37(Suppl
8): S4–S66. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
van Rensburg SJ: Esophageal cancer,
micronutrient malnutrition, and silica fragments. Lancet.
2:1098–1099. 1982. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
National Comprehensive Cancer Network:
NCCN Guidelines and Clinical Resources: Guidelines for Treatment of
Cancer by Site. Version 7. 2015.
|
|
6
|
Brescia AA, Broderick SR, Crabtree TD,
Puri V, Musick JF, Bell JM, Kreisel D, Krupnick AS, Patterson GA
and Meyers BF: Adjuvant therapy for positive nodes after induction
therapy and resection of esophageal cancer. Ann Thorac Surg.
101:200–210. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhang L, Ma J, Han Y, Liu J, Zhou W, Hong
L and Fan D: Targeted therapy in esophageal cancer. Expert Rev
Gastroenterol Hepatol. Feb 19–2016.(Epub ahead of print).
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zeidan Q and Hart GW: The intersections
between O-GlcNAcylation and phosphorylation: Implications for
multiple signaling pathways. J Cell Sci. 123:13–22. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Marshall S, Bacote V and Traxinger RR:
Discovery of a metabolic pathway mediating glucose-induced
desensitization of the glucose transport system. Role of hexosamine
biosynthesis in the induction of insulin resistance. J Biol Chem.
266:4706–4712. 1991.PubMed/NCBI
|
|
10
|
Copeland RJ, Bullen JW and Hart GW:
Cross-talk between GlcNAcylation and phosphorylation: Roles in
insulin resistance and glucose toxicity. Am J Physiol Endocrinol
Metab. 295:E17–E28. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Housley MP, Rodgers JT, Udeshi ND, Kelly
TJ, Shabanowitz J, Hunt DF, Puigserver P and Hart GW: O-GlcNAc
regulates FoxO activation in response to glucose. J Biol Chem.
283:16283–16292. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Vosseller K, Wells L, Lane MD and Hart GW:
Elevated nucleocytoplasmic glycosylation by O-GlcNAc results in
insulin resistance associated with defects in Akt activation in
3T3-L1 adipocytes. Proc Natl Acad Sci USA. 99:5313–5318. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yang JY, Gu JL, Shi JH, Liu F and Shen Q:
The inhibitory effect of OGT gene expression on the level of tau
phosphorylation. Prog Biochem Biophys. 36:346–352. 2009. View Article : Google Scholar
|
|
14
|
Vervoorts J, Lüscher-Firzlaff J and
Lüscher B: The ins and outs of MYC regulation by posttranslational
mechanisms. J Biol Chem. 281:34725–34729. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yang WH, Kim JE, Nam HW, Ju JW, Kim HS,
Kim YS and Cho JW: Modification of p53 with O-linked
N-acetylglucosamine regulates p53 activity and stability. Nat Cell
Biol. 8:1074–1083. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
16
|
Barone BB, Yeh HC, Snyder CF, Peairs KS,
Stein KB, Derr RL, Wolff AC and Brancati FL: Long-term all-cause
mortality in cancer patients with preexisting diabetes mellitus: A
systematic review and meta-analysis. Jama. 300:2754–2764. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Torres CR and Hart GW: Topography and
polypeptide distribution of terminal N-acetylglucosamine residues
on the surfaces of intact lymphocytes. Evidence for O-linked
GlcNAc. J Biol Chem. 259:3308–3317. 1984.PubMed/NCBI
|
|
18
|
Hart GW, Housley MP and Slawson C: Cycling
of O-linked beta-N-acetylglucosamine on nucleocytoplasmic proteins.
Nature. 446:1017–1022. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Qiao Z, Dang C, Zhou B, Li S, Zhang W,
Jiang J, Zhang J, Kong R and Ma Y: O-linked N-acetylglucosamine
transferase (OGT) is overexpressed and promotes O-linked protein
glycosylation in esophageal squamous cell carcinoma. J Biomed Res.
26:268–273. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Love DC and Hanover JA: The hexosamine
signaling pathway: Deciphering the ‘O-GlcNAc code’. Sci STKE.
2005:re132005.PubMed/NCBI
|
|
22
|
Haltiwanger RS, Holt GD and Hart GW:
Enzymatic addition of O-GlcNAc to nuclear and cytoplasmic proteins.
Identification of a uridine diphospho-N-acetylglucosamine: Peptide
beta-N-acetylglucosaminyltransferase. J Biol Chem. 265:2563–2568.
1990.PubMed/NCBI
|
|
23
|
Dong DL and Hart GW: Purification and
characterization of an O-GlcNAc selective
N-acetyl-beta-D-glucosaminidase from rat spleen cytosol. J Biol
Chem. 269:19321–19330. 1994.PubMed/NCBI
|
|
24
|
Cetinbas N, Macauley MS, Stubbs KA,
Drapala R and Vocadlo DJ: Identification of Asp174 and Asp175 as
the key catalytic residues of human O-GlcNAcase by functional
analysis of site-directed mutants. Biochemistry. 45:3835–3844.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Gao Y, Wells L, Comer FI, Parker GJ and
Hart GW: Dynamic O-glycosylation of nuclear and cytosolic proteins:
Cloning and characterization of a neutral, cytosolic
beta-N-acetylglucosaminidase from human brain. J Biol Chem.
276:9838–9845. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Rogacka D, Piwkowska A, Jankowski M,
Kocbuch K, Dominiczak MH, Stepiński JK and Angielski S: Expression
of GFAT1 and OGT in podocytes: Transport of glucosamine and the
implications for glucose uptake into these cells. J Cell Physiol.
225:577–584. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Mi W, Gu Y, Han C, Liu H, Fan Q, Zhang X,
Cong Q and Yu W: O-GlcNAcylation is a novel regulator of lung and
colon cancer malignancy. Biochim Biophys Acta. 1812:514–519. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Caldwell SA, Jackson SR, Shahriari KS,
Lynch TP, Sethi G, Walker S, Vosseller K and Reginato MJ: Nutrient
sensor O-GlcNAc transferase regulates breast cancer tumorigenesis
through targeting of the oncogenic transcription factor FoxM1.
Oncogene. 29:2831–2842. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ngoh GA, Facundo HT, Zafir A and Jones SP:
O-GlcNAc signaling in the cardiovascular system. Circ Res.
107:171–185. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Slawson C, Copeland RJ and Hart GW:
O-GlcNAc signaling: A metabolic link between diabetes and cancer?
Trends Biochem Sci. 35:547–555. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Gu Y, Mi W, Ge Y, Liu H, Fan Q, Han C,
Yang J, Han F, Lu X and Yu W: GlcNAcylation plays an essential role
in breast cancer metastasis. Cancer Res. 70:6344–6351. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Akter H, Park M, Kwon OS, Song EJ, Park WS
and Kang MJ: Activation of matrix metalloproteinase-9 (MMP-9) by
neurotensin promotes cell invasion and migration through ERK
pathway in gastric cancer. Tumour Biol. 36:6053–6062. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Song J, Wu C, Korpos E, Zhang X, Agrawal
SM, Wang Y, Faber C, Schäfers M, Körner H, Opdenakker G, et al:
Focal MMP-2 and MMP-9 activity at the blood-brain barrier promotes
chemokine-induced leukocyte migration. Cell Rep. 10:1040–1054.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Slawson C, Pidala J and Potter R:
Increased N-acetyl-beta-glucosaminidase activity in primary breast
carcinomas corresponds to a decrease in N-acetylglucosamine
containing proteins. Biochim Biophys Acta. 1537:147–157. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Wang YZ, Wu JN, Sun RQ and Wu WX:
Expression of MMP-9 and MMP-9 mRNA in esophageal carcinoma its
correlation with antioncogene p53. Jiang Su Da Xue Xue Bao.
2:113–116. 2005.(In Chinese).
|
|
36
|
Kneass ZT and Marchase RB: Protein
O-GlcNAc modulates motility-associated signaling intermediates in
neutrophils. J Biol Chem. 280:14579–14585. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Hong S, Park KK, Magae J, Ando K, Lee TS,
Kwon TK, Kwak JY, Kim CH and Chang YC: Ascochlorin inhibits matrix
metalloproteinase-9 expression by suppressing activator
protein-1-mediated gene expression through the erk1/2 signaling
pathway: Inhibitory effects of ascochlorin on the invasion of renal
carcinoma cells. J Biol Chem. 280:25202–25209. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Westermarck J and Kähäri VM: Regulation of
matrix metalloproteinase expression in tumor invasion. Faseb J.
13:781–792. 1999.PubMed/NCBI
|