Introduction
Ewing sarcoma (ES) is the second most common primary
bone malignancy, and typically develops in children and
adolescents, predominantly in white males (1). It is also referred to as Ewing sarcoma
family tumor (ESFT), which includes extraskeletal ES and primitive
neuroectodermal tumors. ESFT is a highly aggressive malignancy,
with a rate of metastasis of 27% at the time of diagnosis (1). Chemotherapy with intercalated
locoregional managements, such as surgery and radiation, is the
generally recommended treatment (2).
The advanced development of diagnostic tools and delicate
understanding of transcriptional and translational factors
associated with the pathogenetic Ewing sarcoma breakpoint region 1
(EWSR1)/Friend leukemia integration 1 transcription factor (FLI1)
fusion protein (EWS-FLI1) gene have contributed to the improvement
of targeted therapies for important oncoproteins. Therefore, the
survival rate of patients suffering from ES tends to increase with
better elucidation of the pathogenesis and the application thereof
to the development of management strategies; the 5-year survival
rate may increase from 15 to 39% for metastatic disease, and from
44 to 68% for localized disease (1).
For this reason, it is important to determine and act based upon
the pathognomonic signs of ES; however, the pathogenesis of ES
remains to be elucidated.
There have been a number of suggestions attempting
to explain the complicated pathogenesis of ES, which have included
insulin-like growth factor-binding protein 3 downregulation, sonic
hedgehog signaling and microsatellite-related signaling (3). Additionally, there is evidence to
suggest that expression of the guanosine nucleotide-binding protein
α stimulating (GNAS) gene, which encodes the G-protein α
subunit (Gsα), is associated with the pathogenesis of
ES: i) Insulin-like growth factor 1 receptor (IGF1R)
expression is related to the early growth response 1 (EGR1)
gene and its promoters, which consist of the EWS-FLI1 fusion
protein and the cyclic adenosine monophosphate (cAMP) response
element-binding protein (CREB) (4–6); this
cAMP/CREB signature is activated by Gsα and may result
in EGR1 and IGF1R expression; ii) it has been shown
that CREB-Smad6-Runx2 signaling promotes defective osteogenesis
(7), and the EWS-FLI1 fusion protein
also inhibits Runx2 (8); iii) our
previous genome-wide methylation studies of ES revealed that the
GNAS gene was hypomethylated (9), and proposed that a hypomethylated
GNAS gene may be overexpressed in ES relative to normal
mesenchymal cells of bone, such that ES tumors would have high
expression of Gsα; and iv) activating mutations of the
GNAS gene have been identified in pituitary tumors (10), ovarian granulosa cell tumors (11), renal cell carcinomas (12) and hepatocellular carcinomas (13), and these tumors exhibited
morphological resemblances to neuroendocrine cells, similar to ES.
On the basis of this evidence, we hypothesized that GNAS
expression may be associated with the pathogenesis of ES.
The purposes of the present study were to analyze
GNAS mutation and methylation statuses, and Gsα
expression in ES, in order to determine whether GNAS
expression is pathognomonically relevant. To the best of our
knowledge, the current study is the first to examine the pathogenic
role of the GNAS gene in ES.
Materials and methods
Clinical tumor samples
Formalin-fixed paraffin-embedded (FFPE) tissue
samples from 77 patients with primary ES were obtained at the Kyung
Hee University Hospital in Korea, Central Army Hospital in
Argentina and SARAH Network of Rehabilitation Hospitals in Brazil
between January 2000 and December 2005. Normal control samples were
obtained from the remaining tissues following total knee
replacement surgery due to degenerative osteoarthritis at the Kyung
Hee University Hospital in Korea. At the time of tumor sampling,
patients had no history of chemotherapy or radiation therapy and
there was no evidence of metastatic disease. These tumor samples
were diagnosed according to the World Health Organization criteria
(14) which, in brief, consist of the
following: Small round cell sarcomas, showing diffuse membranous
CD99 immunostaining, cytoplasmic periodic acid-Schiff staining, and
EWSR1 gene translocation demonstrated via fluorescence in
situ hybridization (Zytolight SPEC ROS1 and RET Dual Color
Break Apart Probes; ZytoVision, Bremerhaven, Germany). If an
EWSR1 gene translocation is not identified but a tumor has a
typical immunophenotype in differential diagnosis which is
inconsistent with other small round cell tumors, such as lymphoma
or rhabdomyosarcoma, such tumor samples are diagnosed as ES.
Patient demographics are presented in Table I. Full data, including follow-up
periods and overall survival, were available for 45 patients. The
study protocol was reviewed and approved by the Kyung Hee
University Institutional Review Board (Seoul, South Korea).
 | Table I.Demographics of Ewing sarcoma
patients (n=77). |
Table I.
Demographics of Ewing sarcoma
patients (n=77).
| Clinicopathological
parameter | Value |
|---|
| Age at diagnosis,
years |
|
|
Range | 1–57 |
|
Median | 17 |
| Gender, n (%) |
|
|
Male | 45 (58.4) |
|
Female | 32 (41.6) |
| Tumor site, n
(%) |
|
|
Peripheral | 48 (62.3) |
|
Central | 29 (37.7) |
| Follow-up,
months |
|
|
Range | 6–96 |
|
Median | 30.5 |
| Lung metastasis, n
(%) |
|
|
Present | 6 (7.8) |
|
Absent | 39 (50.6) |
| Not
available | 32 (41.6) |
| Patient outcome, n
(%) |
|
|
Survived | 25 (32.5) |
|
Died | 20 (26.0) |
| Not
available | 32 (41.6) |
Bisulfite conversion and methylation
chip assay
Bisulfite conversions of all DNA samples were
performed using an EZ-96 DNA methylation kit (Zymo Research,
Orange, CA, USA) according to the manufacturer's instructions. For
each bisulfite conversion, 500 ng of genomic DNA was used.
Following bisulfite treatment, quantification of methylcytosine
content was conducted using an Illumina GoldenGate Methylation
Cancer Panel I microarray (Illumina, Inc., San Diego, CA, USA). The
GoldenGate Panel was used to process 1,505 CpG sites from a panel
of 807 cancer-related genes. Briefly, bisulfite-converted DNA was
allowed to react with biotin and was then hybridized to assay
oligos, after which allele-specific extensions and ligations were
conducted at 45°C for 15 min. Ligated products were amplified via
polymerase chain reaction (PCR) with the following parameters: 10
min at 37°C; 34 cycles of 35 sec at 95°C, 35 sec at 56°C and 2 min
at 72°C; 10 min at 72°C; and 5 min at 4°C. Single-stranded PCR
products were prepared by denaturation and hybridized to a Sentrix
Array Matrix (GoldenGate Methylation Cancer Panel I). Array
hybridization was conducted overnight in a temperature gradient
program ranging from 45 to 60°C, and arrays were imaged using a
BeadArray Reader scanner (Illumina, Inc.). Raw methylation ratios
were calculated using the Methylation Module in Illumina's
BeadStudio following background normalization, which was derived by
averaging the signals of a built-in negative control. Each sample
was examined in a duplicate manner in the chip assay.
Direct sequencing
Direct sequencing was performed to detect the
mutational status of GNAS exons 8 and 9. Genomic DNA was
extracted using the Magna Pure LC instrument (Roche Applied
Science, Mannheim, Germany). PCR was performed using a thermal
cycler (GeneAmp PCR system 9700; Thermo Fisher Scientific, Waltham,
MA, USA). PCR ingredients containing 2.5 µl of 10X buffer [50 mM
KCl, 10 mM Tris-HCl (pH 8.3), 15 mM MgCl2 and 0.001%
gelatin], 2.0 µl of 2.5 mM dNTP, 1.0 µl of forward and reverse
primers (10 pmol/µl; Bioneer Corporation, Daejeon, Korea), 0.5 µl
of AmpliTaq® DNA Polymerase (5 U/µl; Thermo Fisher
Scientific) and 1.0 µl of DNA (50 ng/µl) were mixed with deionized
water, at a total volume of 25 µl. PCR conditions for amplifying
GNAS exons 8 and 9 were as follows: Denaturation at 94°C for
2 min; 40 cycles of denaturation at 94°C for 30 sec, annealing at
62°C for 30 sec, and extension at 72°C for 1 min; and a final
extension at 72°C for 10 min. PCR products were purified for
sequencing analysis with the QIAquick PCR purification kit (Qiagen,
Valencia, CA, USA). Cycle sequencing was performed with the
BigDye® Terminator v3.1 Cycle Sequencing Kit (Thermo
Fisher Scientific) according to the manufacturer's protocols, and
the reaction mixture was analyzed on an ABI Prism 3100 DNA
Sequencer (Applied Biosystem; Thermo Fisher Scientific). Primer
sequences for PCR and sequencing are listed in Table II.
 | Table II.Primer sequences for GNAS
exons 8 and 9. |
Table II.
Primer sequences for GNAS
exons 8 and 9.
|
| Primer
sequence |
|
|---|
|
|
|
|
|---|
| GNAS
exon | Forward | Reverse | Amplicon size
(bp) |
|---|
| 8 | 5′-GTT TCG GTT GGC
TTT GGT GA-3′ | 5′-TGG CTT ACT GGA
AGT TGA CT-3′ | 129 |
| 9 | 5′-GAC ATT CAC CCC
AGT CCC TCT-3′ | 5′-GAA GCA AAG CGT
TCT TTA CGA-3′ | 155 |
Immunohistochemistry
Immunohistochemical stains for anti-G protein α S
antibody were performed on FFPE human specimens.
Immunohistochemistry procedures were performed on 5 µm tissue
sections on a Leica Bond-Max automatic slide stainer (Leica
Biosystems Melbourne Pty. Ltd., Melbourne, Australia) using the
standard protocol. In brief, the 5 µm sections of FFPE tissues were
deparaffinized using Bond Dewax Solution (Cat#. AR9222; Leica
Biosystems Newcastle Ltd.), and an antigen retrieval procedure was
performed using Proteinase K Solution (ready for use; Cat#. S3020;
Dako Korea Co., Ltd., Seoul, Korea) for 7 min at room tempurature.
The endogenous peroxidase was quenched by incubation with hydrogen
peroxide for 15 min. Sections were incubated for 15 min at ambient
temperate with a rabbit monoclonal anti-G protein α S antibody
(Cat#. ab83735; Abcam, Cambridge, MA, USA) at a 1:600 dilution.
Secondary antibodies goat anti-rabbit biotin-free polymeric
horseradish peroxidase and rabbit anti-mouse linker antibody,
contained in a Bond Polymer Refine Detection System (Cat#. DS9800;
Leica Biosystems Newcastle Ltd., Newcastle, UK), were incubated for
8 min at room temperature. Bound primary antibodies were visualized
using DAB with a Bond Polymer Refine Detection System (Cat#.
DS9800; Leica Biosystems Newcastle Ltd.) and a Bond-Max automatic
slide stainer (Leica Biosystems Melbourne Pty. Ltd.). The nuclei in
these sections were counterstained with hematoxylin using a Bond
Polymer detection System (Cat#. 9800; Leica Biosystems Newcastle,
Ltd.). Pancreatic islet cells, obtained from the remaining tissues
of patients that received pancreatectomies due to chronic
pancreatitis, were used as a positive external control.
Pathological analysis of
immunohistochemistry
The immunohistochemical results were measured and
scored with respect to intensity and proportion in positive tumor
cells, and were independently reviewed by three pathologists (Drs
Byeong-Joo Noh, Ji-Youn Sung and Yong-Koo Park). The staining
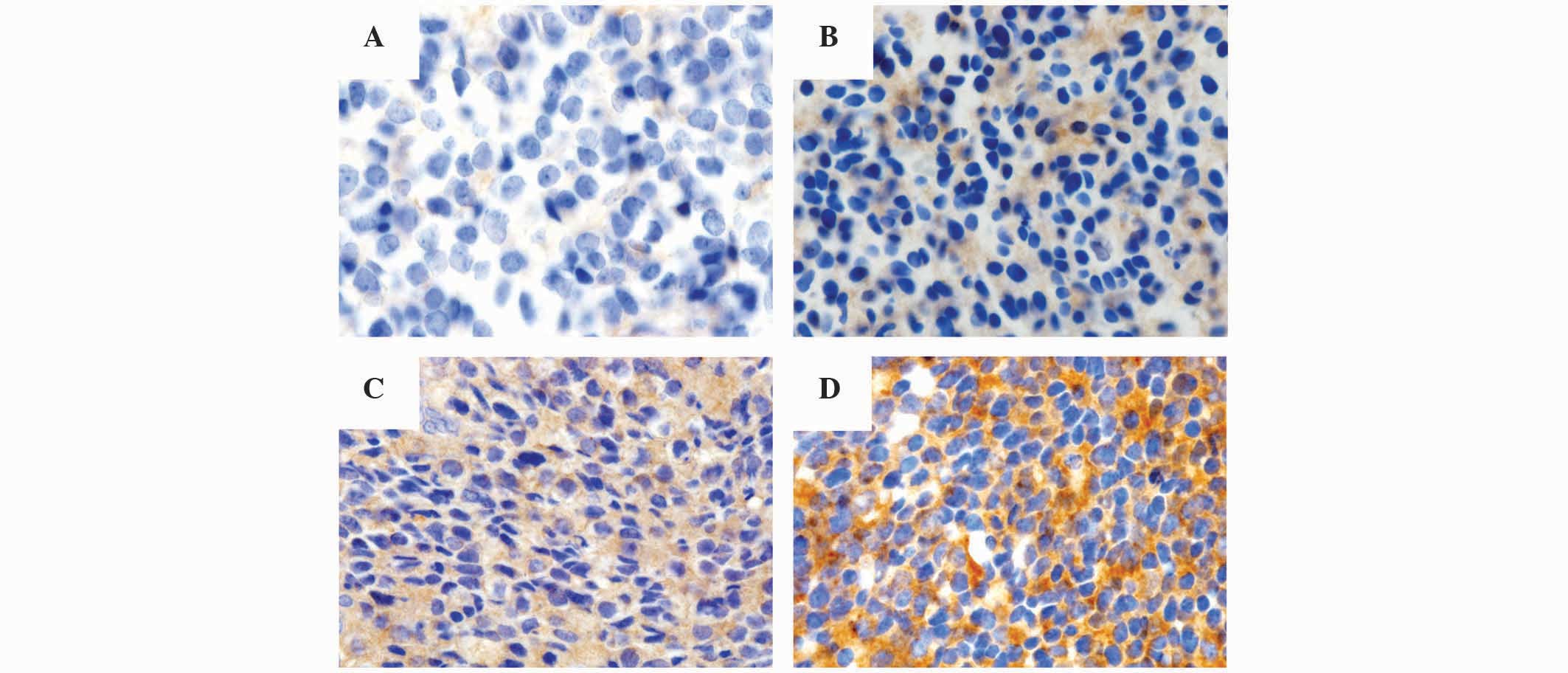
intensity was graded in a 4-tiered system as follows: No visible
brown staining, 0; pale brown, 1+; non-homogeneous brown, 2+;
homogeneous dark brown color, 3+. According to the cytoplasmic
staining intensity and proportion of positive tumor cells, the
final scores of Gsα were categorized as grade 0, 1, 2 or
3: Grade 0, absence of Gsα staining in 100% of tumor
cells; grade 1, intensity 1+ in >70% of tumor cells or intensity
2+ in ≤30% of tumor cells; grade 2, intensity 1+ in >70% of
tumor cells, intensity 2+ in >30% but ≤70% of tumor cells or
intensity 3+ ≤30% of tumor cells; grade 3, intensity 2+ in >70%
of tumor cells or intensity of 3+ in >30% of tumor cells
(Fig. 1).
Statistical analysis
Statistical analyses were conducted using SPSS
version 12.0 (SPSS, Inc., Chicago, IL, USA). Pearson's
χ2 and independent t-tests were conducted to determine
correlations between tested values and clinicopathological
parameters. Univariate survival analyses were performed to examine
the prognostic significance of antibody expression and
clinicopathological parameters, according to the Kaplan-Meier curve
with a log-rank test. Statistical significance was considered to be
indicated by P<0.05.
Results
Methylation analysis of the GNAS
gene
The degree of methylation of the GNAS gene
was assessed using the Illumina GoldenGate Methylation Cancer Panel
I microarray. The GoldenGate DNA methylation method measures DNA
methylation levels as β-values ranging from 0 (no DNA methylation
detected) to 1 (complete DNA methylation). The results indicated
that the GNAS gene in ES tumor samples was less methylated
than in normal controls; the mean β-value was 0.48 in ES tumor
samples vs. 0.83 in normal control samples, indicating that the
GNAS gene was overexpressed in ES relative to normal
tissue.
Mutation analysis of the GNAS
gene
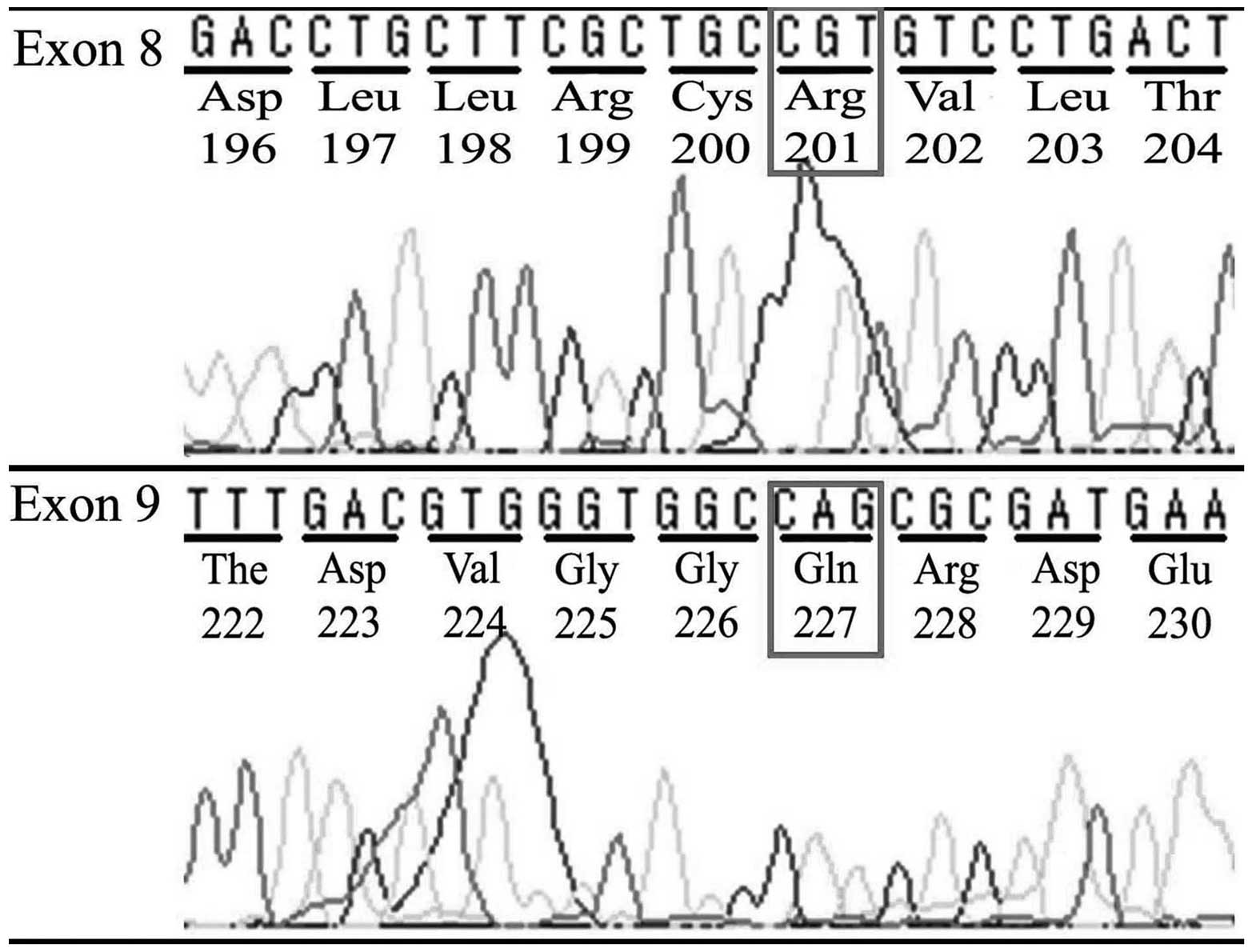
No mutations were detected in exons 8 or 9 of the
GNAS locus complex on chromosome 20q13.3 in DNA extracted
from any of the FFPE tumor samples from the ES patients (Fig. 2), demonstrating that the pathogenesis
of ES was not associated with GNAS mutation.
Immunohistochemical analysis of
Gsα expression
The correlation of Gsα expression with
clinicopathological parameters was analyzed using a binary system
approach, grouping low expression (grades 0–1) vs. high expression
(grades 2–3). Clinicopathological cases involving missing values or
without available clinical information were abbreviated for
statistical analyses. Of the 52 ES tumor samples, 34 samples
(65.4%) showed high Gsα expression, compared with 18
sample (34.6%) with low Gsα expression.
Gsα expression correlated well with the
methylation status of the GNAS gene; β-values were
0.681±0.304 in tumor samples with low expression of Gsα,
compared with 0.245±0.229 in samples with high Gsα
expression (P=0.001) (Table III).
High Gsα expression in tumor samples was found in 14/17
samples (82.4%) with a hypomethylated GNAS gene vs. 1/5
samples (20%) with a hypermethylated GNAS gene (P=0.009;
Table III).
 | Table III.Association of clinicopathological
parameters with Gsα expression. |
Table III.
Association of clinicopathological
parameters with Gsα expression.
|
| Gsα
expression |
|
|
|---|
|
|
|
|
|
|---|
| Clinicopathological
parametera | Low | High | Total | P-value |
|---|
| Age, mean ± SD | 21.17±11.03 | 18.62±10.60 | 19.16 ± 10.50 | 0.420b |
| Gender, n (%) |
|
|
| 0.727c |
|
Male | 12 (36.4) | 21 (63.6) | 33 (100.0) |
|
|
Female | 6
(31.6) | 13 (68.4) | 19 (100.0) |
|
| Site involved, n
(%) |
|
|
| 0.451c |
|
Peripheral | 13 (38.2) | 21 (61.8) | 34 (100.0) |
|
|
Central | 5
(27.8) | 13 (72.2) | 18 (100.0) |
|
| β-value, mean ±
SD | 0.681±0.304 | 0.245±0.229 | 0.478 ± 0.345 | 0.001b |
| Degree of
methylation, n (%) |
|
|
| 0.009c |
|
Hypomethylation | 3
(17.6) | 14 (82.4) | 17 (100.0) |
|
|
Hypermethylation | 4
(80.0) | 1
(20.0) | 5
(100.0) |
|
| Outcome, n (%) |
|
|
| 0.055c |
|
Survived | 2
(13.3) | 13 (86.7) | 15 (100.0) |
|
|
Died | 6
(46.2) | 7
(53.8) | 13 (100.0) |
|
Notably, high Gsα expression was detected
more frequently in samples from living patients than decedents,
although this was not statistically significant; high
Gsα was found in 12/15 samples (86.7%) from living
patients, vs. 7/13 samples (53.8%) from decedents (P=0.055;
Table III). Furthermore,
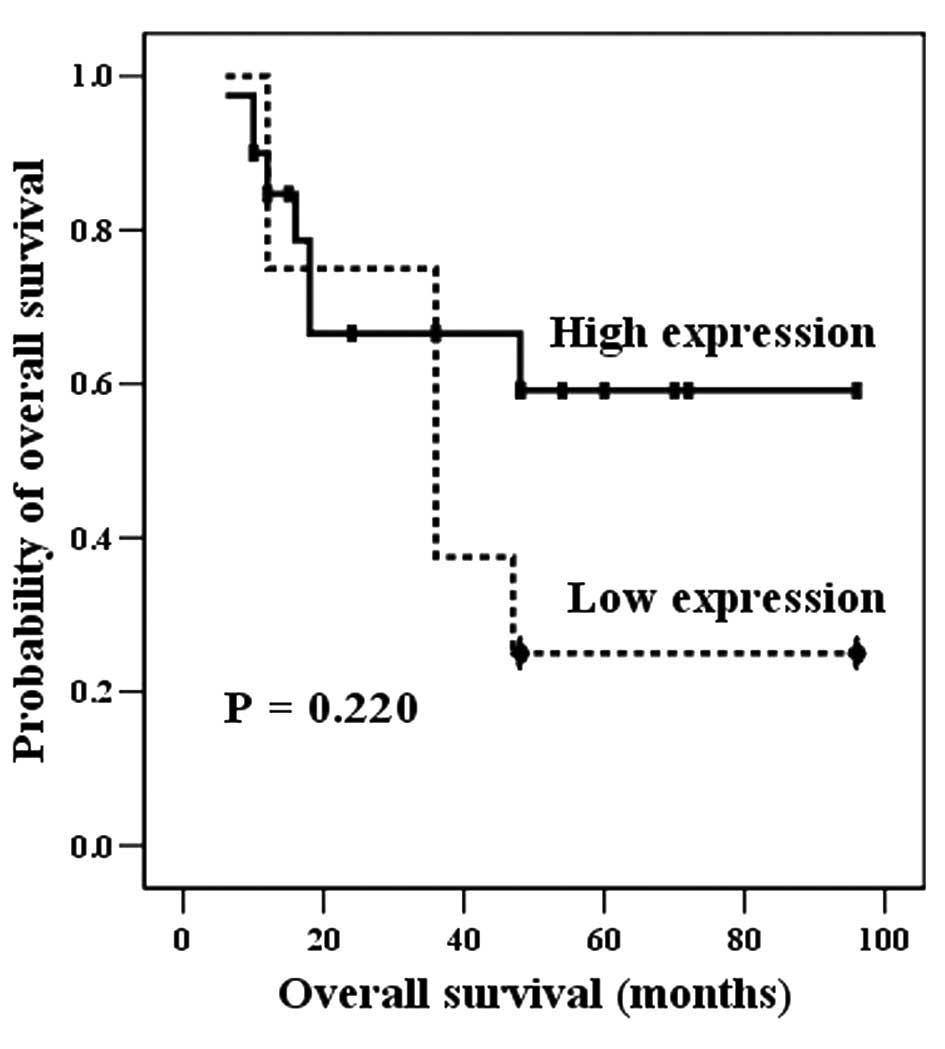
Gsα levels were not found to significantly correlate
with the survival rate (P=0.220; Fig.
3).
Discussion
The GNAS gene on chromosome 20q13.3 encodes
the α subunit of the heterotrimeric G protein complex
(Gsα) (15). Activating or
inactivating mutations and epigenetic changes at the GNAS
locus have been described in a variety of human diseases (15).
Activating mutations of the GNAS gene induce
protein alterations as follows: A mutation at exon 8 of the
GNAS gene is responsible for substitution of arginine at
codon 201 with cysteine or histidine, termed R201C or R201H,
respectively; and, more rarely, a mutation at exon 9 of GNAS
results in substitution of the glutamine at codon 227 with leucine,
arginine, lysine or histidine, termed Q227L, Q227R, Q227K or Q227H,
respectively (16). These protein
alterations may inhibit GTPase activity, maintaining an active form
of Gsα. Activating mutations of the GNAS locus
have been detected in McCune-Albright syndrome (17), pituitary adenoma (10), ovarian granulosa cell tumor (11), renal cell carcinoma (12), hepatocellular carcinoma (13), pancreatic intestinal-type intraductal
papillary mucinous neoplasm (18,19) and
myelodysplastic syndrome (20). As
with mesenchymal bone tumors, fibrous dysplasia (2,21) and
parosteal osteosarcoma (22) have
also been linked to activating mutations of the GNAS gene.
It has been documented that GNAS status has diagnostic
utility for differentiating fibro-osseous lesions in
morphologically challenging diagnoses: Fibrous dysplasia vs.
ossifying fibroma, adamantinoma or osteofibrous dysplasia (16,22,23).
GNAS mutation was not identified in any ES tumor sample in
the current study, indicating that GNAS mutation is not
associated with the pathogenesis of ES. For this reason,
GNAS mutation analysis may be used to exclude metastatic
bone lesions with GNAS mutations, including renal cell
carcinoma or hepatocellular carcinoma, which exhibit morphological
resemblances to ES tumor cells (24,25).
Inactivating mutations of the GNAS locus from
the maternal or paternal germ-line are attributed to amino acid
substitutions, nonsense mutations, inversions, splicing site
mutations, insertions or deletions. Progressive osseous heterotopia
has been verified to result from inactivating GNAS mutations
of predominantly paternal origin (21,26). It
has also been documented that epigenetic alterations of the
GNAS gene are associated with deletion of the syntaxin 16
gene and hypomethylation of the A/B domain at the GNAS locus
complex (27). Epigenetic changes in
the GNAS gene have been demonstrated to be important in
disease progression in pseudohypoparathyroidism type Ib (28). Until now, there has been no research
demonstrating that activating or inactivating mutations or
epigenetic changes of GNAS have prognostic and not
diagnostic value. The T393C pleomorphism of the GNAS locus
has been identified to have prognostic value in various malignant
tumors, including clear cell renal cell carcinoma (29), bladder cancer (30), colorectal cancer (31), epithelial ovarian cancer (32), melanoma (33), glioblastoma (34) and non-small cell lung cancer (35).
In the current study, the epigenetic methylation
status of the GNAS gene in ES tumor samples was
significantly associated with Gsα expression: The less
methylated GNAS gene in ES tumor samples overexpressed
Gsα (Table III).
However, ES patients with hypomethylated GNAS genes (high
expression of Gsα) had increased survival probability
relative to those with hypermethylated GNAS genes (low
expression of Gsα), with a non-significant positive
trend (P=0.055, Table III; P=0.220,
Fig. 3). On the basis of these
results, we speculate that the epigenetic transformation of the
GNAS gene to hypermethylation status (low Gsα
expression) in ES tumors may have a an association with more
aggressive behavior of ES tumors, but is not significantly
correlated with the overall survival.
In summary, GNAS mutation is not associated
with the pathogenesis of ES tumors. This finding may be used to
distinguish metastatic bone lesions with GNAS mutations that
have morphological similarities to ES tumors. Analysis of the
methylation status of the GNAS gene and immunohistochemical
Gsα expression suggests that hypermethylated GNAS
gene (low Gsα expression) in ES may be associated with
unfavorable progression with a non-significant trend. Further
studies with a larger sample of patients are required to verify
these results.
References
|
1
|
Esiashvili N, Goodman M and Marcus RB Jr:
Changes in incidence and survival of Ewing sarcoma patients over
the past 3 decades: Surveillance epidemiology and end results data.
J Pediatr Hematol Oncol. 30:425–430. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Lee SE, Lee EH, Park H, Sung JY, Lee HW,
Kang SY, Seo S, Kim BH, Lee H, Seo AN, et al: The diagnostic
utility of the GNAS mutation in patients with fibrous dysplasia:
Meta-analysis of 168 sporadic cases. Hum Pathol. 43:1234–1242.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Lessnick SL and Ladanyi M: Molecular
pathogenesis of Ewing sarcoma: New therapeutic and transcriptional
targets. Annu Rev Pathol. 7:145–159. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ma Y, Cheng Q, Ren Z, Xu L, Zhao Y, Sun J,
Hu S and Xiao W: Induction of IGF-1R expression by EGR-1
facilitates the growth of prostate cancer cells. Cancer Lett.
317:150–156. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wu X, Cheng J, Li P, Yang M, Qiu S, Liu P
and Du J: Mechano-sensitive transcriptional factor Egr-1 regulates
insulin-like growth factor-1 receptor expression and contributes to
neointima formation in vein grafts. Arterioscler Thromb Vasc Biol.
30:471–476. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Watson DK, Robinson L, Hodge DR, Kola I,
Papas TS and Seth A: FLI1 and EWS-FLI1 function as ternary complex
factors and ELK1 and SAP1a function as ternary and quaternary
complex factors on the Egr1 promoter serum response elements.
Oncogene. 14:213–221. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Fan QM, Yue B, Bian ZY, Xu WT, Tu B, Dai
KR, Li G and Tang TT: The CREB-Smad6-Runx2 axis contributes to the
impaired osteogenesis potential of bone marrow stromal cells in
fibrous dysplasia of bone. J Pathol. 228:45–55. 2012.PubMed/NCBI
|
|
8
|
Li X, McGee-Lawrence ME, Decker M and
Westendorf JJ: The Ewing's sarcoma fusion protein, EWS-FLI, binds
Runx2 and blocks osteoblast differentiation. J Cell Biochem.
111:933–943. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Park HR, Jung WW, Kim HS and Park YK:
Microarray-based DNA methylation study of Ewing's sarcoma of the
bone. Oncol Lett. 8:1613–1617. 2014.PubMed/NCBI
|
|
10
|
Gadelha MR, Trivellin G, Hernández,
Ramírez LC and Korbonits M: Genetics of pituitary adenomas. Front
Horm Res. 41:111–140. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kalfa N, Ecochard A, Patte C, Duvillard P,
Audran F, Pienkowski C, Thibaud E, Brauner R, Lecointre C, Plantaz
D, et al: Activating mutations of the stimulatory g protein in
juvenile ovarian granulosa cell tumors: A new prognostic factor? J
Clin Endocrinol Metab. 91:1842–1847. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kalfa N, Lumbroso S, Boulle N, Guiter J,
Soustelle L, Costa P, Chapuis H, Baldet P and Sultan C: Activating
mutations of Gsalpha in kidney cancer. J Urol. 176:891–895. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Nault JC, Fabre M, Couchy G, Pilati C,
Jeannot E, Van Tran Nhieu J, Saint-Paul MC, De Muret A, Redon MJ,
Buffet C, et al: GNAS-activating mutations define a rare subgroup
of inflammatory liver tumors characterized by STAT3 activation. J
Hepatol. 56:184–191. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
de alava E, Lessnick SL and Sorensen PH:
Ewing sarcoma. WHO classification of Tumours of Soft Tissue and
Bone. Fletcher CDM, Bridge JA, Hogendoorn PCW and Mertens F: (4th).
IARC Press. (Lyon). 305–309. 2012.
|
|
15
|
Weinstein LS, Liu J, Sakamoto A, Xie T and
Chen M: Minireview: GNAS: Normal and abnormal functions.
Endocrinology. 145:5459–5464. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Tabareau-Delalande F, Collin C,
Gomez-Brouchet A, Decouvelaere AV, Bouvier C, Larousserie F, Marie
B, Delfour C, Aubert S, Rosset P, et al: Diagnostic value of
investigating GNAS mutations in fibro-osseous lesions: A
retrospective study of 91 cases of fibrous dysplasia and 40 other
fibro-osseous lesions. Mod Pathol. 26:911–921. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Salpea P and Stratakis CA: Carney complex
and McCune Albright syndrome: An overview of clinical
manifestations and human molecular genetics. Mol Cell Endocrinol.
386:85–91. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Matthaei H, Wu J, Dal Molin M, Shi C,
Perner S, Kristiansen G, Lingohr P, Kalff JC, Wolfgang CL, Kinzler
KW, et al: GNAS sequencing identifies IPMN-specific mutations in a
subgroup of diminutive pancreatic cysts referred to as ‘incipient
IPMNs’. Am J Surg Pathol. 38:360–363. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Dal Molin M, Matthaei H, Wu J, Blackford
A, Debeljak M, Rezaee N, Wolfgang CL, Butturini G, Salvia R, Bassi
C, et al: Clinicopathological correlates of activating GNAS
mutations in intraductal papillary mucinous neoplasm (IPMN) of the
pancreas. Ann Surg Oncol. 20:3802–3808. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Bejar R, Stevenson K, Abdel-Wahab O,
Galili N, Nilsson B, Garcia-Manero G, Kantarjian H, Raza A, Levine
RL, Neuberg D and Ebert BL: Clinical effect of point mutations in
myelodysplastic syndromes. N Engl J Med. 364:2496–2506. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Regard JB, Malhotra D, Gvozdenovic-Jeremic
J, Josey M, Chen M, Weinstein LS, Lu J, Shore EM, Kaplan FS and
Yang Y: Activation of hedgehog signaling by loss of GNAS causes
heterotopic ossification. Nat Med. 19:1505–1512. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Carter JM, Inwards CY, Jin L, Evers B,
Wenger DE, Oliveira AM and Fritchie KJ: Activating GNAS mutations
in parosteal osteosarcoma. Am J Surg Pathol. 38:402–409. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Shi RR, Li XF, Zhang R, Chen Y and Li TJ:
GNAS mutational analysis in differentiating fibrous dysplasia and
ossifying fibroma of the jaw. Mod Pathol. 26:1023–1031. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kalfa N, Lumbroso S, Boulle N, Guiter J,
Soustelle L, Costa P, Chapuis H, Baldet P and Sultan C: Activating
mutations of Gsalpha in kidney cancer. J Urol. 176:891–895. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Nault JC, Fabre M, Couchy G, Pilati C,
Jeannot E, Van Nhieu Tran J, Saint-Paul MC, De Muret A, Redon MJ,
Buffet C, et al: GNAS-activating mutations define a rare subgroup
of inflammatory liver tumors characterized by STAT3 activation. J
Hepatol. 56:184–191. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Pignolo RJ, Xu M, Russell E, Richardson A,
Kaplan J, Billings PC, Kaplan FS and Shore EM: Heterozygous
inactivation of Gnas in adipose-derived mesenchymal progenitor
cells enhances osteoblast differentiation and promotes heterotopic
ossification. J Bone Miner Res. 26:2647–2655. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Turan S, Ignatius J, Moilanen JS, Kuismin
O, Stewart H, Mann NP, Linglart A, Bastepe M and Jüppner H: De novo
STX16 deletions: An infrequent cause of pseudohypoparathyroidism
type Ib that should be excluded in sporadic cases. J Clin
Endocrinol Metab. 97:E2314–E2319. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Yuno A, Usui T, Yambe Y, Higashi K, Ugi S,
Shinoda J, Mashio Y and Shimatsu A: Genetic and epigenetic states
of the GNAS complex in pseudohypoparathyroidism type Ib using
methylation-specific multiplex ligation-dependent probe
amplification assay. Eur J Endocrinol. 168:169–175. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Frey UH, Lümmen G, Jäger T, Jöckel KH,
Schmid KW, Rübben H, Müller N, Siffert W and Eisenhardt A: The
GNAS1 T393C polymorphism predicts survival in patients with clear
cell renal cell carcinoma. Clin Cancer Res. 12:759–763. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Frey UH, Eisenhardt A, Lümmen G, Rübben H,
Jöckel KH, Schmid KW and Siffert W: The T393C polymorphism of the G
alpha s gene (GNAS1) is a novel prognostic marker in bladder
cancer. Cancer Epidemiol Biomarkers Prev. 14:871–877. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Frey UH, Alakus H, Wohlschlaeger J,
Schmitz KJ, Winde G, van Calker HG, Jöckel KH, Siffert W and Schmid
KW: GNAS1 T393C polymorphism and survival in patients with sporadic
colorectal cancer. Clin Cancer Res. 11:5071–5077. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Tominaga E, Tsuda H, Arao T, Nishimura S,
Takano M, Kataoka F, Nomura H, Hirasawa A, Aoki D and Nishio K:
Amplification of GNAS may be an independent, qualitative and
reproducible biomarker to predict progression-free survival in
epithelial ovarian cancer. Gynecol Oncol. 118:160–166. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Frey UH, Fritz A, Rotterdam S, Schmid KW,
Potthoff A, Altmeyer P, Siffert W and Brockmeyer NH: GNAS1 T393C
polymorphism and disease progression in patients with malignant
melanoma. Eur J Med Res. 15:422–427. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
El Hindy N, Lambertz N, Bachmann HS, Frey
UH, Adamzik M, Zhu Y, Sure U, Siffert W and Sandalcioglu IE: Role
of the GNAS1 T393C polymorphism in patients with glioblastoma
multiforme. J Clin Neurosci. 18:1495–1499. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Xie FJ, Zhao P, Kou JY, Hong W, Fu L, Hu
L, Hong D, Su D, Gao Y and Zhang YP: The T393C polymorphism of
GNAS1 as a predictor for chemotherapy sensitivity and survival in
advanced non-small-cell lung cancer patients treated with
gemcitabine plus platinum. Cancer Chemother Pharmacol.
69:1443–1448. 2012. View Article : Google Scholar : PubMed/NCBI
|