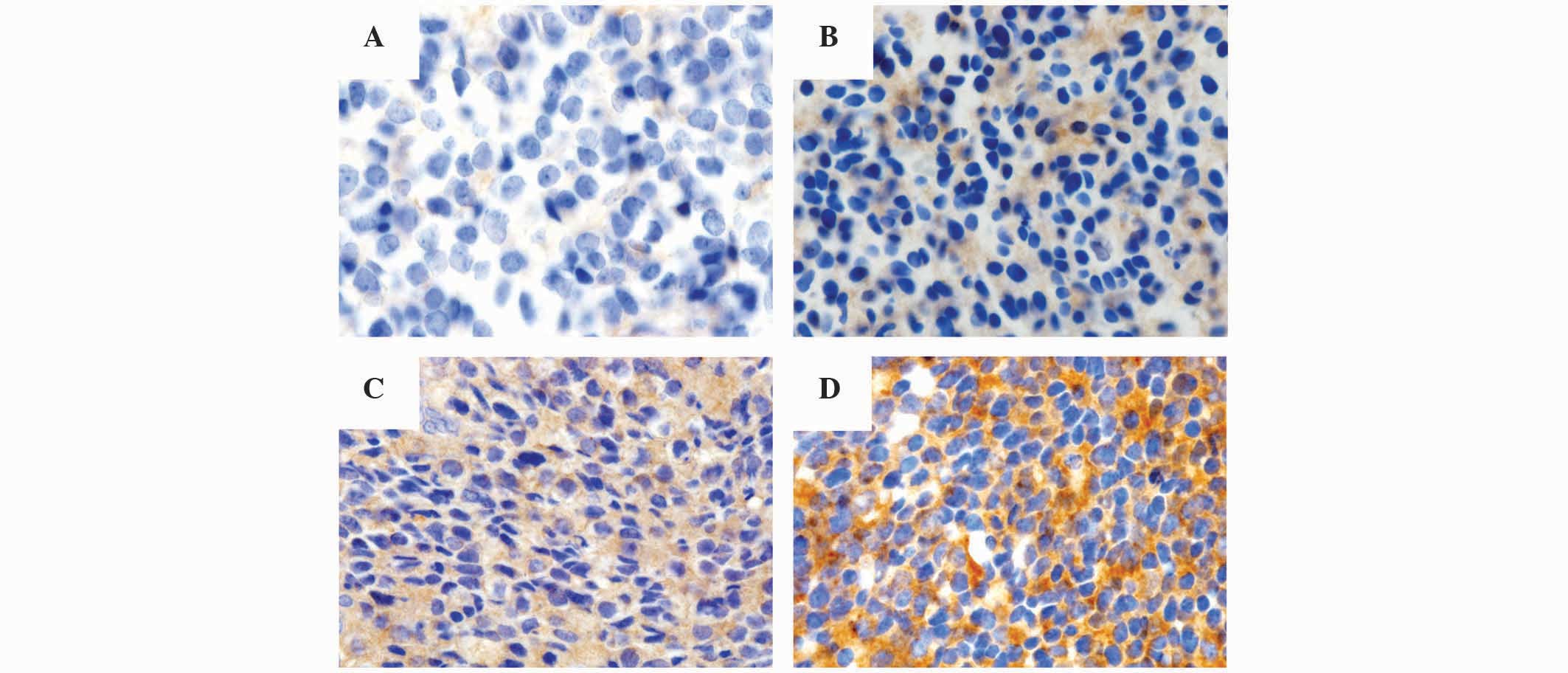
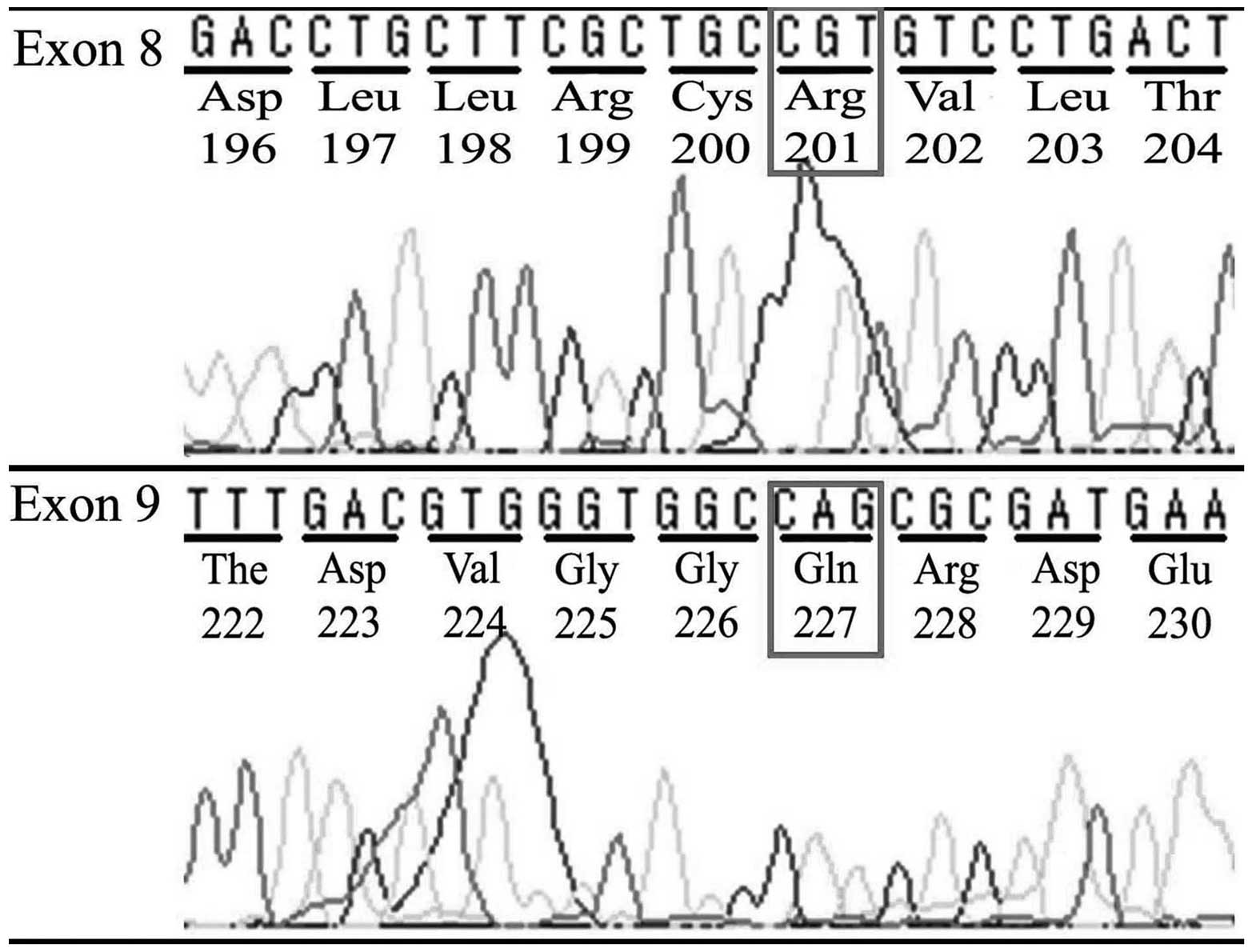
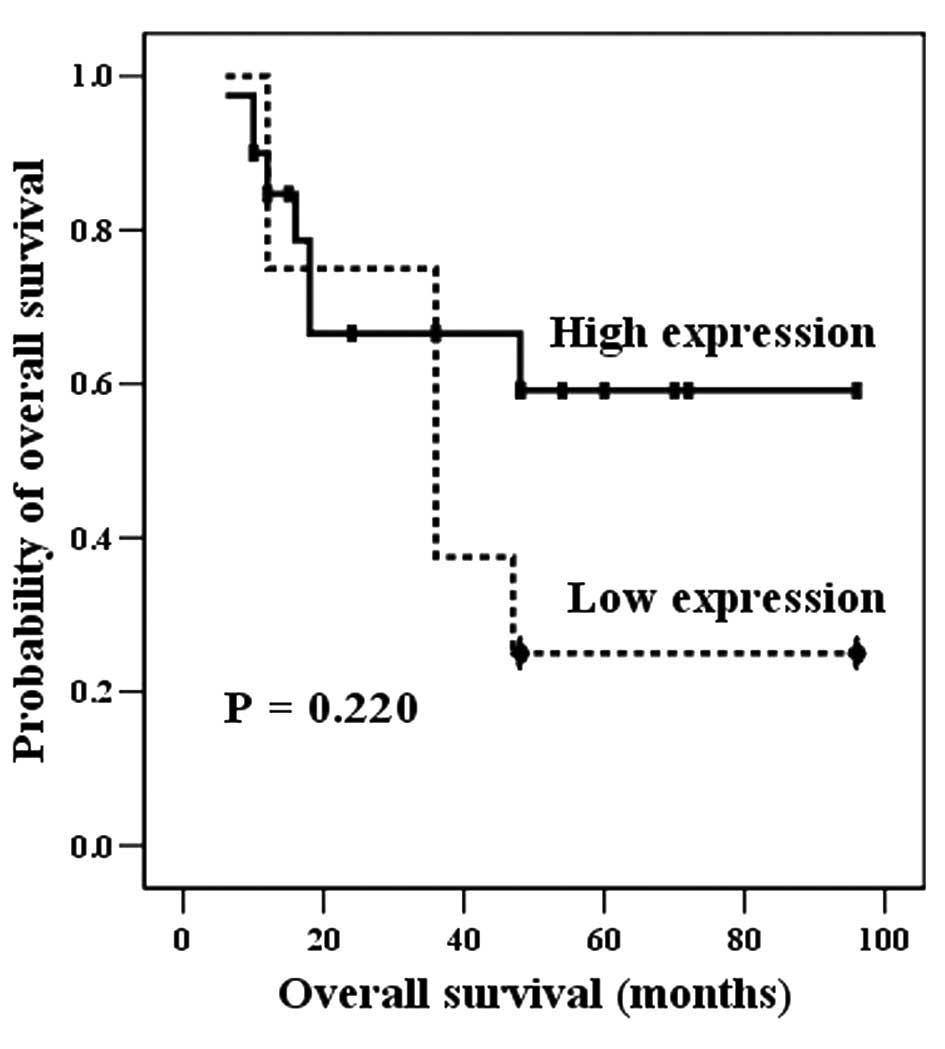
|
1
|
Esiashvili N, Goodman M and Marcus RB Jr:
Changes in incidence and survival of Ewing sarcoma patients over
the past 3 decades: Surveillance epidemiology and end results data.
J Pediatr Hematol Oncol. 30:425–430. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Lee SE, Lee EH, Park H, Sung JY, Lee HW,
Kang SY, Seo S, Kim BH, Lee H, Seo AN, et al: The diagnostic
utility of the GNAS mutation in patients with fibrous dysplasia:
Meta-analysis of 168 sporadic cases. Hum Pathol. 43:1234–1242.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Lessnick SL and Ladanyi M: Molecular
pathogenesis of Ewing sarcoma: New therapeutic and transcriptional
targets. Annu Rev Pathol. 7:145–159. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ma Y, Cheng Q, Ren Z, Xu L, Zhao Y, Sun J,
Hu S and Xiao W: Induction of IGF-1R expression by EGR-1
facilitates the growth of prostate cancer cells. Cancer Lett.
317:150–156. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wu X, Cheng J, Li P, Yang M, Qiu S, Liu P
and Du J: Mechano-sensitive transcriptional factor Egr-1 regulates
insulin-like growth factor-1 receptor expression and contributes to
neointima formation in vein grafts. Arterioscler Thromb Vasc Biol.
30:471–476. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Watson DK, Robinson L, Hodge DR, Kola I,
Papas TS and Seth A: FLI1 and EWS-FLI1 function as ternary complex
factors and ELK1 and SAP1a function as ternary and quaternary
complex factors on the Egr1 promoter serum response elements.
Oncogene. 14:213–221. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Fan QM, Yue B, Bian ZY, Xu WT, Tu B, Dai
KR, Li G and Tang TT: The CREB-Smad6-Runx2 axis contributes to the
impaired osteogenesis potential of bone marrow stromal cells in
fibrous dysplasia of bone. J Pathol. 228:45–55. 2012.PubMed/NCBI
|
|
8
|
Li X, McGee-Lawrence ME, Decker M and
Westendorf JJ: The Ewing's sarcoma fusion protein, EWS-FLI, binds
Runx2 and blocks osteoblast differentiation. J Cell Biochem.
111:933–943. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Park HR, Jung WW, Kim HS and Park YK:
Microarray-based DNA methylation study of Ewing's sarcoma of the
bone. Oncol Lett. 8:1613–1617. 2014.PubMed/NCBI
|
|
10
|
Gadelha MR, Trivellin G, Hernández,
Ramírez LC and Korbonits M: Genetics of pituitary adenomas. Front
Horm Res. 41:111–140. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kalfa N, Ecochard A, Patte C, Duvillard P,
Audran F, Pienkowski C, Thibaud E, Brauner R, Lecointre C, Plantaz
D, et al: Activating mutations of the stimulatory g protein in
juvenile ovarian granulosa cell tumors: A new prognostic factor? J
Clin Endocrinol Metab. 91:1842–1847. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kalfa N, Lumbroso S, Boulle N, Guiter J,
Soustelle L, Costa P, Chapuis H, Baldet P and Sultan C: Activating
mutations of Gsalpha in kidney cancer. J Urol. 176:891–895. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Nault JC, Fabre M, Couchy G, Pilati C,
Jeannot E, Van Tran Nhieu J, Saint-Paul MC, De Muret A, Redon MJ,
Buffet C, et al: GNAS-activating mutations define a rare subgroup
of inflammatory liver tumors characterized by STAT3 activation. J
Hepatol. 56:184–191. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
de alava E, Lessnick SL and Sorensen PH:
Ewing sarcoma. WHO classification of Tumours of Soft Tissue and
Bone. Fletcher CDM, Bridge JA, Hogendoorn PCW and Mertens F: (4th).
IARC Press. (Lyon). 305–309. 2012.
|
|
15
|
Weinstein LS, Liu J, Sakamoto A, Xie T and
Chen M: Minireview: GNAS: Normal and abnormal functions.
Endocrinology. 145:5459–5464. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Tabareau-Delalande F, Collin C,
Gomez-Brouchet A, Decouvelaere AV, Bouvier C, Larousserie F, Marie
B, Delfour C, Aubert S, Rosset P, et al: Diagnostic value of
investigating GNAS mutations in fibro-osseous lesions: A
retrospective study of 91 cases of fibrous dysplasia and 40 other
fibro-osseous lesions. Mod Pathol. 26:911–921. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Salpea P and Stratakis CA: Carney complex
and McCune Albright syndrome: An overview of clinical
manifestations and human molecular genetics. Mol Cell Endocrinol.
386:85–91. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Matthaei H, Wu J, Dal Molin M, Shi C,
Perner S, Kristiansen G, Lingohr P, Kalff JC, Wolfgang CL, Kinzler
KW, et al: GNAS sequencing identifies IPMN-specific mutations in a
subgroup of diminutive pancreatic cysts referred to as ‘incipient
IPMNs’. Am J Surg Pathol. 38:360–363. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Dal Molin M, Matthaei H, Wu J, Blackford
A, Debeljak M, Rezaee N, Wolfgang CL, Butturini G, Salvia R, Bassi
C, et al: Clinicopathological correlates of activating GNAS
mutations in intraductal papillary mucinous neoplasm (IPMN) of the
pancreas. Ann Surg Oncol. 20:3802–3808. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Bejar R, Stevenson K, Abdel-Wahab O,
Galili N, Nilsson B, Garcia-Manero G, Kantarjian H, Raza A, Levine
RL, Neuberg D and Ebert BL: Clinical effect of point mutations in
myelodysplastic syndromes. N Engl J Med. 364:2496–2506. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Regard JB, Malhotra D, Gvozdenovic-Jeremic
J, Josey M, Chen M, Weinstein LS, Lu J, Shore EM, Kaplan FS and
Yang Y: Activation of hedgehog signaling by loss of GNAS causes
heterotopic ossification. Nat Med. 19:1505–1512. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Carter JM, Inwards CY, Jin L, Evers B,
Wenger DE, Oliveira AM and Fritchie KJ: Activating GNAS mutations
in parosteal osteosarcoma. Am J Surg Pathol. 38:402–409. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Shi RR, Li XF, Zhang R, Chen Y and Li TJ:
GNAS mutational analysis in differentiating fibrous dysplasia and
ossifying fibroma of the jaw. Mod Pathol. 26:1023–1031. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kalfa N, Lumbroso S, Boulle N, Guiter J,
Soustelle L, Costa P, Chapuis H, Baldet P and Sultan C: Activating
mutations of Gsalpha in kidney cancer. J Urol. 176:891–895. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Nault JC, Fabre M, Couchy G, Pilati C,
Jeannot E, Van Nhieu Tran J, Saint-Paul MC, De Muret A, Redon MJ,
Buffet C, et al: GNAS-activating mutations define a rare subgroup
of inflammatory liver tumors characterized by STAT3 activation. J
Hepatol. 56:184–191. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Pignolo RJ, Xu M, Russell E, Richardson A,
Kaplan J, Billings PC, Kaplan FS and Shore EM: Heterozygous
inactivation of Gnas in adipose-derived mesenchymal progenitor
cells enhances osteoblast differentiation and promotes heterotopic
ossification. J Bone Miner Res. 26:2647–2655. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Turan S, Ignatius J, Moilanen JS, Kuismin
O, Stewart H, Mann NP, Linglart A, Bastepe M and Jüppner H: De novo
STX16 deletions: An infrequent cause of pseudohypoparathyroidism
type Ib that should be excluded in sporadic cases. J Clin
Endocrinol Metab. 97:E2314–E2319. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Yuno A, Usui T, Yambe Y, Higashi K, Ugi S,
Shinoda J, Mashio Y and Shimatsu A: Genetic and epigenetic states
of the GNAS complex in pseudohypoparathyroidism type Ib using
methylation-specific multiplex ligation-dependent probe
amplification assay. Eur J Endocrinol. 168:169–175. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Frey UH, Lümmen G, Jäger T, Jöckel KH,
Schmid KW, Rübben H, Müller N, Siffert W and Eisenhardt A: The
GNAS1 T393C polymorphism predicts survival in patients with clear
cell renal cell carcinoma. Clin Cancer Res. 12:759–763. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Frey UH, Eisenhardt A, Lümmen G, Rübben H,
Jöckel KH, Schmid KW and Siffert W: The T393C polymorphism of the G
alpha s gene (GNAS1) is a novel prognostic marker in bladder
cancer. Cancer Epidemiol Biomarkers Prev. 14:871–877. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Frey UH, Alakus H, Wohlschlaeger J,
Schmitz KJ, Winde G, van Calker HG, Jöckel KH, Siffert W and Schmid
KW: GNAS1 T393C polymorphism and survival in patients with sporadic
colorectal cancer. Clin Cancer Res. 11:5071–5077. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Tominaga E, Tsuda H, Arao T, Nishimura S,
Takano M, Kataoka F, Nomura H, Hirasawa A, Aoki D and Nishio K:
Amplification of GNAS may be an independent, qualitative and
reproducible biomarker to predict progression-free survival in
epithelial ovarian cancer. Gynecol Oncol. 118:160–166. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Frey UH, Fritz A, Rotterdam S, Schmid KW,
Potthoff A, Altmeyer P, Siffert W and Brockmeyer NH: GNAS1 T393C
polymorphism and disease progression in patients with malignant
melanoma. Eur J Med Res. 15:422–427. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
El Hindy N, Lambertz N, Bachmann HS, Frey
UH, Adamzik M, Zhu Y, Sure U, Siffert W and Sandalcioglu IE: Role
of the GNAS1 T393C polymorphism in patients with glioblastoma
multiforme. J Clin Neurosci. 18:1495–1499. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Xie FJ, Zhao P, Kou JY, Hong W, Fu L, Hu
L, Hong D, Su D, Gao Y and Zhang YP: The T393C polymorphism of
GNAS1 as a predictor for chemotherapy sensitivity and survival in
advanced non-small-cell lung cancer patients treated with
gemcitabine plus platinum. Cancer Chemother Pharmacol.
69:1443–1448. 2012. View Article : Google Scholar : PubMed/NCBI
|