Introduction
Triple-negative breast cancer (TNBC), an aggressive
type of breast cancer, lacks effective targeted therapy due to the
absence of hormone receptors and human epidermal growth factor-2
(HER2). Therefore, considerable effort has been made to identify
subclasses of TNBC with distinct characteristics that may
potentially be targeted in the clinic.
In 2008, Cheang et al (1) revealed that TNBC cases that positively
expressed epidermal growth factor receptor (EGFR) or cytokeratin
(CK) 5/6 demonstrated a shorter survival time and poorer response
to chemotherapy, but might benefit from EGFR-targeted therapy
(2–7).
Another marker in TNBC with potential prognostic and therapeutic
value, androgen receptor (AR), has drawn particular attention since
2010 (8). In recent years, studies
have progressed to the molecular level. Prat et al (9) investigated the correlation between TNBC
molecular subtypes and the PAM50 intrinsic subtypes as well as the
claudin-low subtype. These authors observed that the majority of
TNBCs were either basal-like (39 to 54%) or claudin-low (25% to
39%), followed by HER2-enriched and luminal. However, Lehmann et
al (10) reported another
classification based on gene expression profiles of 587 TNBCs:
basal-like 1 (BL1), basal-like 2 (BL2), immunomodulatory (IM),
mesenchymal (M), mesenchymal stem-like (MSL), and luminal androgen
receptor (LAR). Further analysis narrowed these down to three main
groups (BL, mesenchymal-like and LAR), which demonstrated different
responses to cytotoxic and targeted therapies.
These apparently different classifications may be
related (11). Basal-like in the
PAM50 assay encompassed the TNBC BL subtypes defined by Lehmann as
well as certain tumors classified as IM and M (10,12). In
addition, MSL describes a similar group of claudin-low cancers
while LAR shares a number of gene expression features of estrogen
receptor (ER)-positive and HER2-enriched cancers (10,12). Thus,
despite the lack of consensus, it appears reasonable to predict
that there are three basic subtypes within TNBC (11,13–15).
Gene expression-based classification significantly
changes our understanding of the heterogeneity of TNBC. However, it
raises the question of how this sophisticated approach can be
translated into a practical and clinically accessible diagnostic
test, given that gene identification is currently not feasible for
large-scale application on routine formalin-fixed paraffin-embedded
clinical samples (16). In this
study, we adopt the immunohistochemistry (IHC) methodology. We
examined the IHC profile of 154 TNBC cases and identified three
subtypes exhibiting diverse clinicopathological and prognostic
characteristics with the minimum use of biomarkers.
Patients and methods
Patient selection
We collected breast cancer cases with sufficient
medical records from the Department of Breast Surgery, China-Japan
Union Hospital of Jilin University, China, between January 2006 and
November 2014. Inclusion criteria for this study were: i) female;
ii) primary stage I–III invasive breast cancer; iii) no neoadjuvant
chemotherapy or radiotherapy prior to surgery; iv) breast tissue
samples available for study. All of the subjects underwent surgical
treatment according to standard treatment protocols.
Clinicopathological parameters including age, histological subtype,
tumor size, histological grade, nodal status, and presence of
lymphovascular invasion and tumor necrosis were noted. The
histological subtype and histological grade were assessed in
accordance with standard guidelines and confirmed independently by
two pathologists from the Department of Pathology at the
China-Japan Union Hospital of Jilin University. The median
follow-up time was 68 months (range, 2 to 108 months). The study
was approved by the ethics committee of Jilin University.
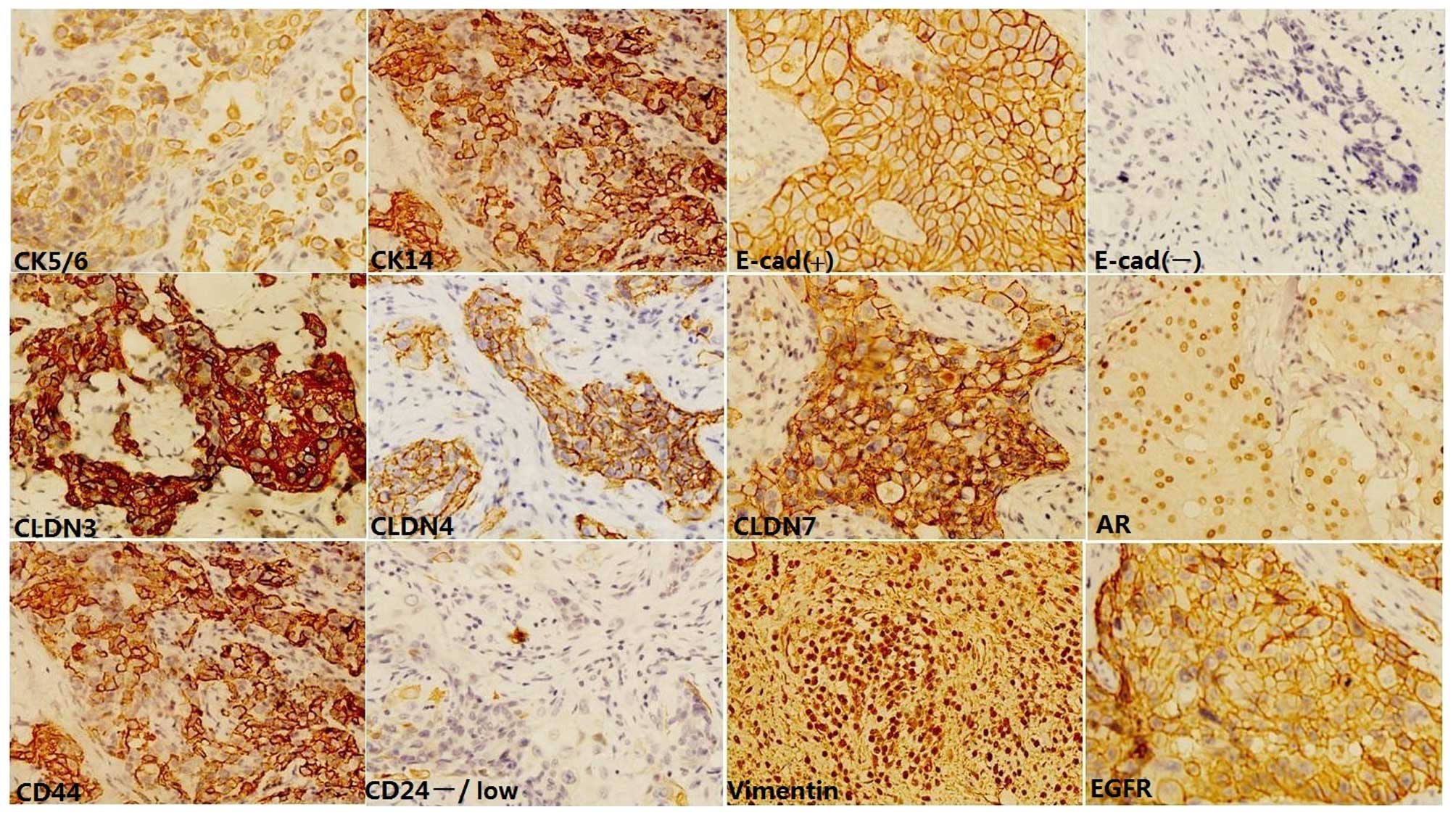
Immunohistochemistry and scoring
Immunohistochemical staining was performed according
to the following protocol. Sections from paraffin-embedded tissue
microarrays were cut to 4 µm, deparaffinized in xylene and
rehydrated through graded alcohols. Microwave epitope retrieval was
performed in target retrieval pH 6.0 (Dako, Carpinteria, CA, USA)
for ER and HER2, high pH target retrieval for CK5/6 (Dako), or 10
mM citrate buffer (pH 6.0) for 10 min followed by cooling for 15
min at room temperature for claudins.
The following antibodies were used: clone SP1
against ER (1:300 dilution; Dako), clone SP2 against progesterone
receptor (PR; 1:250 dilution; Neomarkers, Fremont, CA, USA), clone
SP3 against HER2 (1:200 dilution; Neomarkers), clone SP6 against
Ki67 (1:200 dilution; Neomarkers), clone D5/16B4 against CK5/6
(1:100 dilution; M7237; Dako), clone LL002 against CK14 (1:20;
NCL-LL002; Novocastra, Newcastle upon Tyne, UK), clone E30 against
EGFR (1:50; M7239; Dako), clone NCH-38 against E-cadherin (1:50,
Dako), clone V9 against vimentin (1:150, Dako), clone Z23.JM
against claudin 3 (1:300; Invitrogen Life Technologies, Carlsbad,
CA, USA), clone Ab15104 against claudin 4 (1:300, Abcam), clone
Ab27287 against claudin 7 (1:400; Abcam, Cambridge, MA), clone AR27
against AR (1:100, NCL-AR-318), clone 156-3C11 against CD44 (1:100,
Cell Signaling Technology, Inc., Danvers, MA, USA), and clone
Ab2-SN3b against CD24 (1:100 Neomarkers).
Staining results were assessed by two pathologists
in a blinded fashion. For ER, PR and AR status, stains were
considered positive if at least 1% of tumor nuclei demonstrated
positivity, regardless of the intensity (1 to 3+). For HER2 status,
stains were considered positive if at least 30% of tumor cells
exhibited a cell membrane staining score of 3+. There are no
commonly accepted cut-off points reported for EGFR. Membranous EGFR
staining in >1% of tumor cells was used as the definition of
protein positivity according to the Dako criteria provided in the
pharmDx kit instructions. For Ki67, the mean percentage of nuclear
positivity was evaluated in a stepwise manner; i.e. 1, 2, 3, 5, 10,
15, 20, 25, 30, 35, 40, 50, 60, 70, 80 and 90%. For CK5/6 and CK14,
staining was scored as positive when more than 10% of the tumor
cells demonstrated cytoplasmic and/or membranous staining.
E-cadherin expression was analyzed semi-quantitatively according to
the percentage of cells demonstrating membrane positivity: 0,
0–10%; 1+, 10–30%; 2+, 30–70%; 3+, > 70%. E-cadherin expression
was considered positive when scores were ≥2 and negative when
scores were ≤1. Any distinct positive staining of the tumor
cytoplasm in cancer cells with the vimentin antibody was regarded
as positive vimentin expression. Claudin immunoreactivity was
assessed based on a combined score of the extension and intensity
of membrane expression. The extension was registered as the
percentage of positive cells for claudins: 0, 0%; 1+, <25%; 2+,
25–50%; 3+, >50%. The intensity of membrane immunostaining was
graded as: 0 (negative); 1 (weak); 2 (moderate); 3 (strong). The
two scores were multiplied to give an overall score of 0–9, of
which 0 was considered negative, 1–2 was considered weak, 3–6
moderate, and 9 strong staining. Negative and weak expression was
considered as low, while moderate and strong were considered high.
Tumors with low expression of all three claudins were defined as
claudin-low. For CD44 and CD24, stains were scored positive when
more than 10% of the tumor cells exhibited membranous staining. We
considered a tumor to have a cancer stem cell (CSC) phenotype when
the frequency of CD44+CD24-/low cells was more than 10%, as
previously described in other studies (17,18). Any
discordant scores were reviewed together by the two scorers to
obtain a consensus.
Definition of breast cancer subtypes
by IHC
Identifying subgroups of TNBC is of significance for
a better understanding of this complex disease. By drawing on the
work of Prat et al (9,13) and Lehmann et al (10,12) on
gene expression subtypes, we attempted for the first time to
classify TNBC into three subsets using IHC markers. Our main aim
was to seek IHC surrogates that potentially identify the main three
gene expression subtypes. Here are certain noteworthy points from
the studies of Prat et al and Lehmann et al: i) TNBC
subtypes defined by Lehmann differentially correlate with the PAM50
intrinsic subtypes (12,13). BL1, BL2, IM and M cases are primarily
composed of the BL intrinsic subtype (99%, 95%, 84% and 97%,
respectively), while ~50% of MSL cases and none of LAR cases have
the BL intrinsic subtype (12).
Therefore, the vast majority of non-basal TNBCs are MSL and LAR
tumors. In addition, BL1, BL2, IM and M subtypes express higher
levels of basal cytokeratin expression (i.e., CK5/6 and CK14),
while tumors in the MSL category exhibit significantly lower basal
cytokeratin expression and LAR tumors lack basal cytokeratin
expression (10,12). ii) LAR shares a number of gene
expression features of ER+ and HER2-enriched cancers
(10). AR protein is highly expressed
within the LAR subgroup, on average >10-fold higher than all
other subtypes (10). iii) MSL is
characterized by enrichment for gene expression patterns associated
with epithelial-to-mesenchymal (ETM) transition (10,12,19). A
portion of the MSL subtypes also are enriched for the CSC-like
phenotype (10,12,19), and
exhibit low expression of tight junction proteins including claudin
3, 4 and 7 (10,12,19),
consistent with a group of cancers previously described as
claudin-low (9). iv) The three main
subtypes (BL, mesenchymal-like and LAR) defined by Lehmann et
al (10) are concordant with the
three main groups previously identified by Prat et al (BL,
claudin-low and luminal/HER2-enriched) (13) and by Neve et al (20) and Kao et al (21), based upon cell lines alone (basal A,
associated with the ETS pathway and BRCA1 signatures and resembling
BL tumors; basal B, exhibiting mesenchymal and stem/progenitor cell
characteristics; and luminal, exhibiting an ER signature and
resembling luminal A/B tumors).
Together, it appears feasible to translate the gene
expression subtypes into three IHC subtypes. Based on the first
point above, triple-negative cases which also positively express
either CK5/6 or CK14 are referred to as the ‘BL’ group in this
article. Therefore, the BL subgroup in this article likely
encompasses the BL1, BL2, IM and M subtypes and a small proportion
of the MSL tumors that express basal cytokeratin. Based on the
second point, triple-negative cases which also positively express
AR are referred to as the AR+ group. However, selecting IHC marker
panels to define the third group is relatively challenging.
According to the classification defined by Lehmann et al,
the majority of the third group consists of MSL tumors that lack
basal cytokeratin expression (12),
whereas according to Prat et al (13), the third group should be claudin-low.
Although MSL and claudin-low share certain similar features, they
are not synonymous. All MSL tumors are associated with EMT
transition (10,11), which is characterized by
downregulation of E-cadherin and occludin and induction of
mesenchymal marker proteins including vimentin and fibronectin
(22–25), while only a portion of MSL cases are
claudin-low, enriched in CSC-like features with an absence of
claudin proteins. Therefore, in order to distinguish the most
appropriate IHC surrogates for the third group, we explored the ETM
phenotype (evaluating vimentin and E-cadherin expression), CSC-like
phenotype (analyzing CD44 and CD24 expression), and claudin 3, 4
and 7 expression in all triple-negative cases, and then defined the
third group as vimentin+ and E-cadherin-; CD44+CD24-/low
phenotype; low expression of all three claudins, respectively.
Vimentin and E-cadherin are well-established and widely accepted as
markers for EMT (20–23), while CD44+CD24-/low is a known marker
for the CSC-like phenotype (9,21,26,27).
Statistical analysis
Differences in the frequencies of basic
characteristics and clinicopathological parameters among breast
cancer subtypes were examined using Chi-square tests, or Fisher's
exact test in the case of less than five expected cases.
Relapse-free survival (RFS) was defined as the time from the date
of diagnosis to the date of relapse of breast cancer, including
locoregional recurrence and/or distant metastasis. Breast
cancer-specific survival (BCSS) was defined as the date of a
patient's diagnosis of breast cancer until mortality. Survival
times were censored if the primary or underlying cause of mortality
was not breast cancer, or if the patient was still alive on
December 30, 2014 (the date when the outcome data were collected).
Survival curves were obtained using the Kaplan-Meier method and
differences in survival among the breast cancer subtypes were
assessed by the log-rank test. Prognostic analyses used the Cox
regression method. Univariate analyses tested classical
clinicopathological features: age (>50 vs. ≤50), pathological
tumor size (pT2-3 vs. pT1), lymph node status (positive vs.
negative), histological grade (2 or 3 vs. 1), necrosis (marked vs.
minimal or absent), Ki67 (>30% vs. ≤30%), adjuvant chemotherapy
(performed vs. not performed). The findings were analyzed using
SPSS statistical software for Windows, version 18 (SPSS, Inc.,
Chicago, IL, USA. All statistical tests were two-sided, and
P<0.05 was considered to indicate a statistically significant
difference.
Results
Patient characteristics
There were a total of 2407 breast cancer patients
receiving surgery at the China-Japan Union Hospital of Jilin
University between January 2006 and November 2014. Among these,
1646 cases that had informative IHC results were included in the
study. The median age at diagnosis in the study population was 54
years (range, 23–87 years). Mastectomy was performed in 78.3% of
cases (1289/1646), and 21.7% (357/1646) underwent breast conserving
surgery. Following surgery, 82.6% (1360/1646) received adjuvant
chemotherapy. The remaining 286 (17.4%) patients did not receive
any adjuvant systemic chemotherapy. The median follow-up time was
68 months (range, 2 to 108 months). Of the 1646 patients, 154 had
triple-negative breast cancer (TNBC). The clinicopathological
characteristics and IHC profiles of the TNBC cases and other types
of breast cancer (non-TNBC) are presented in Table I. The Chi-square test revealed a
statistically significant difference in tumor size, histological
grade, tumor necrosis and Ki67 labeling index between TNBC and
non-TNBC patients. The two groups also differed in the levels of
AR, CK5/6, CK14, EGFR, E-cadherin, vimentin, claudin 3, 4 and 7
expression and CD44+CD24-/low phenotype (Fig. 1). TNBCs had a statistically larger
percentage of tumors that were positive for CK5/6 (57.8%), CK14
(39.6%), EGFR (59.0%), vimentin (44.2%) and CD44+CD24-/low
phenotype (27.3%) compared with non-TNBCs (2.2%, 2.1%, 6.8%, 7.2%
and 2.4%, respectively). AR, E-cadherin, and claudins 3, 4 and 7
staining was greater in non-TNBCs (83.2%, 71.7%, 97.6%, 97.2% and
97.4%, respectively), whereas the positivity for these five markers
in TNBCs was 11.7%, 43.5%, 68.2%, 74.0% and 72.7% (P=0.000).
 | Table I.Clinicopathological characteristics
of TNBC and non-TNBC patients. |
Table I.
Clinicopathological characteristics
of TNBC and non-TNBC patients.
|
Characteristics | TNBC n=154 | Non-TNBC
n=1492 | P-value |
|---|
| Age |
|
| 0.649 |
|
≤50 | 85 |
852 |
|
|
>50 | 69 |
640 |
|
| Family history of
breast cancer |
|
| 0.131 |
| No | 141 | 1411 |
|
|
Yes | 13 |
81 |
|
| Histological
type |
|
|
Invasive ductal carcinoma | 93 | 1082 |
|
|
Invasive lobular
carcinoma | 5 |
143 |
|
|
Medullary carcinoma | 13 |
82 |
|
|
Metaplastic carcinoma | 12 |
78 |
|
|
Apocrine carcinoma | 13 |
78 |
|
|
Others | 6 |
29 |
|
| Pathological tumor
size |
|
| 0.002 |
|
pT1 | 58 |
792 |
|
|
pT2-3 | 96 |
700 |
|
| Histological
grade |
|
| <0.001 |
| 1 | 23 |
228 |
|
| 2 | 34 |
943 |
|
| 3 | 97 |
321 |
|
| Pathological
axillary lymph |
|
| 0.326 |
| node status |
|
Negative | 91 |
820 |
|
|
Positive | 63 |
672 |
|
| Lymphovascular
invasion |
|
| 0.707 |
|
Absent | 116 | 1103 |
|
|
Present | 38 |
389 |
|
| Necrosis |
|
| <0.001 |
| Minimal
or absent | 89 | 1368 |
|
|
Marked | 65 |
124 |
|
| Ki67 |
|
| <0.001 |
|
≤30% | 72 | 1125 |
|
|
>30% | 82 |
367 |
|
| AR |
|
| <0.001 |
|
Negative | 136 |
251 |
|
|
Positive | 18 | 1241 |
|
|
CK5/6 |
|
| <0.001 |
|
Negative | 65 | 1459 |
|
|
Positive | 89 | 33 |
|
| CK14 |
|
| <0.001 |
|
Negative | 93 | 1461 |
|
|
Positive | 61 | 31 |
|
| EGFR |
|
| <0.001 |
|
Negative | 94 | 1390 |
|
|
Positive | 60 |
102 |
|
| E-cadherin |
|
| <0.001 |
|
Negative | 87 |
422 |
|
|
Positive | 67 | 1070 |
|
| Vimentin |
|
| <0.001 |
|
Negative | 86 | 1385 |
|
|
Positive | 68 |
107 |
|
| Claudin 3 |
|
| <0.001 |
|
Negative | 49 | 36 |
|
|
Positive | 105 | 1456 |
|
| Claudin 4 |
|
| <0.001 |
|
Negative | 40 | 42 |
|
|
Positive | 114 | 1450 |
|
| Claudin 7 |
|
| <0.001 |
|
Negative | 42 |
39 |
|
|
Positive | 112 | 1453 |
|
|
CD44+CD24−/low |
|
| <0.001 |
| No | 112 | 1456 |
|
|
Yes | 42 |
36 |
|
| RFS event |
|
|
|
| No | 104 | 1208 |
|
|
Yes | 50 |
284 |
|
| Chemotherapy |
|
|
|
| No | 31 |
255 |
|
|
Yes | 123 | 1237 |
|
| Mean survival
time | 86.3 | 98.7 |
|
| (95%
CI) | (79.7–93.1) | (81.2–114.5) |
|
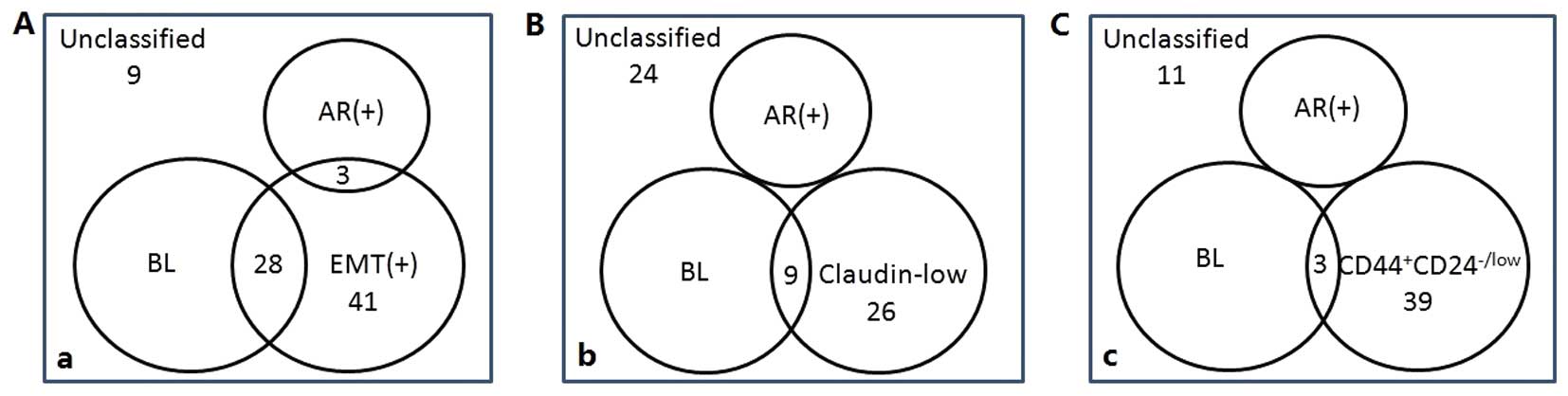
New IHC classification of TNBC
As described in the Patients and methods section, we
defined triple-negative cases which also positively expressed
either CK5/6 or CK14 as the BL group, triple-negative cases which
also positively expressed AR as the AR+ group, and respectively
defined the third group as vimentin+ and E-cadherin-;
CD44+CD24-/low phenotype; low expression of claudins 3,
4 and 7. A comparison of these three different classifications is
shown in Fig. 2. A lower level of
overlap was observed between the BL group and the third group when
the third group was defined as CD44+CD24-/low phenotype, and the
proportion of unclassified cases was also relatively smaller in
this classification model. Therefore, the three subtypes of TNBC
designated in this study are the BL group (83 cases), AR+ group (18
cases), and CD44+CD24-/low phenotype (39 cases). Eleven
cases that were unclassified and three cases that overlapped
between the BL group and CD44+CD24-/low phenotype were excluded in
the following study.
The clinicopathological characteristics of each TNBC
subtype are shown in Table II. When
a difference among the three groups was detected, multiple
comparison was carried out to assess where the difference lies. The
Chi-square test revealed that the three subcategories exhibited
significantly different characteristics in terms of age, tumor
size, histological grade, presence/absence of tumor necrosis and
Ki67 labeling index. Multiple comparison further demonstrated that
the three subtypes differed significantly from each other in
histological grade and tumor necrosis, but not in age, tumor size
or Ki67 labeling index. The histological grade of the
CD44+CD24-/low subtype was often grade 3 (53.8%) or
grade 2 (28.2%), which was lower than tumors in the BL group (grade
3, 81.9%; grade 2, 14.4%), and higher than those in the AR+ group
(grade 3, 16.7%; grade 2, 33.3%). A total of 38.5% of
CD44+CD24-/low subtype cases demonstrated marked tumor necrosis, a
percentage intermediate between that of the BL group (57.8%) and
the AR+ group (11.1%).
 | Table II.Clinicopathological characteristics
of triple-negative breast cancer immunohistochemical subtypes. |
Table II.
Clinicopathological characteristics
of triple-negative breast cancer immunohistochemical subtypes.
|
Characteristics | Basal-like
n=83 | AR+n=18 |
CD44+CD24−/low
n=39 | P-value |
|---|
| Age |
|
|
| 0.038 |
|
≤50 | 52 |
6a | 18 |
|
|
>0 | 31 | 12 | 21 |
|
| Family history of
breast cancer |
|
|
| 0.839 |
| No | 76 | 17 | 35 |
|
|
Yes | 7 | 1 | 4 |
|
| Histological
type |
|
|
|
|
|
Invasive ductal carcinoma | 63 | 6 | 14 |
|
|
Invasive lobular
carcinoma | 2 | 1 | 1 |
|
|
Medullary carcinoma | 11 | 0 | 2 |
|
|
Metaplastic carcinoma | 3 | 0 | 19 |
|
|
Apocrine carcinoma | 2 | 11 | 0 |
|
|
Others | 2 | 0 | 3 |
|
| Pathological tumor
size |
|
|
| 0.041 |
|
pT1 | 24 | 10a | 18 |
|
|
pT2-3 | 59 | 8 | 21 |
|
| Histological
grade |
|
|
| <0.001 |
| 1 | 3 |
9a,b |
7a,b |
|
| 2 | 12 | 6 | 11 |
|
| 3 | 68 | 3 | 21 |
|
| Pathological
axillary lymph node status |
|
|
| 0.734 |
|
Negative | 47 | 12 | 23 |
|
|
Positive | 36 | 6 | 16 |
|
| Lymphovascular
invasion |
|
|
| 0.174 |
|
Absent | 63 | 16 | 26 |
|
|
Present | 20 | 2 | 13 |
|
| Necrosis |
|
|
| <0.001 |
| Minimal
or absent | 35 | 16a,b | 24a,b |
|
|
Marked | 48 | 2 | 15 |
|
| Ki67 |
|
|
| 0.003 |
|
≤30% | 29 | 14a,b | 19a,b |
|
|
<30% | 54 | 4 | 20 |
|
| EGFR |
|
|
| 0.006 |
|
Negative | 34 | 8 | 30a,b |
|
|
Positive | 41 | 6 | 9 |
|
| E-cadherin |
|
|
| <0.001 |
|
Negative | 38 | 4 | 37a,b |
|
|
Positive | 45 | 14 | 2 |
|
| Vimentin |
|
|
| <0.001 |
|
Negative | 55 | 15 |
4a,b |
|
|
Positive | 28 | 3 | 35 |
|
| Claudin 3 |
|
|
| <0.001 |
|
Negative | 10 | 2 | 36a,b |
|
|
Positive | 73 | 16 | 3 |
|
| Claudin 4 |
|
|
| <0.001 |
|
Negative | 9 | 1 |
29a,b |
|
|
Positive | 74 | 17 | 10 |
|
| Claudin 7 |
|
|
| <0.001 |
|
Negative | 11 | 1 |
28a,b |
|
|
Positive | 72 | 17 | 11 |
|
| Chemotherapy |
|
|
|
0.512 |
| No | 14 | 4 | 10 |
|
|
Yes | 69 | 14 | 29 |
|
| RFS event |
|
|
|
|
| No | 51 | 15 | 26 |
|
|
Yes | 32 | 3 | 13 |
|
| Mean survival
time | 75.8 | 96.3 | 84.7 |
|
| (95%
CI) | (59.9–88.4) | (84.0–105.7) | (73.4–94.2) |
|
As for age and tumor size, although the Chi-square
test revealed a statistically significant difference among the
three subcategories, multiple comparison revealed that only the
difference between the BL group and AR+ group was significant.
Patients with AR+ tumors were older than patients with BL tumors
(>50 years, 66.7% vs. 37.3%; multiple comparison test,
P=0.0226). A total of 55.5% of AR+ tumors measured ≤2 cm (pT1)
while 28.9% of BL tumors were pT1 (multiple comparison test,
P=0.0301). In the multiple comparison test, although the
CD44+CD24-/low subtype did not reveal distinct
characteristics in age and tumor size when separately compared with
the BL group and AR+ group, the percentage of patients older than
50 years (53.8%) and the percentage of pT1 tumors (46.2%) were
intermediate between the BL group and AR+ group.
As for the Ki67 labeling index, multiple comparison
revealed that a significant difference existed between AR+ group
and BL group (P<0.001), and also between AR+ group
and CD44+CD24−/low group (P=0.0389). However,
BL group and CD44+CD24−/low group did not
differ in Ki67 (P=0.1463). A total of 22.2% of AR+ tumors had a
Ki67 labeling index >30%, which indicated a less proliferative
subtype compared with the BL group (65.1%) and CD44+CD24-/low
subtype (51.3%).
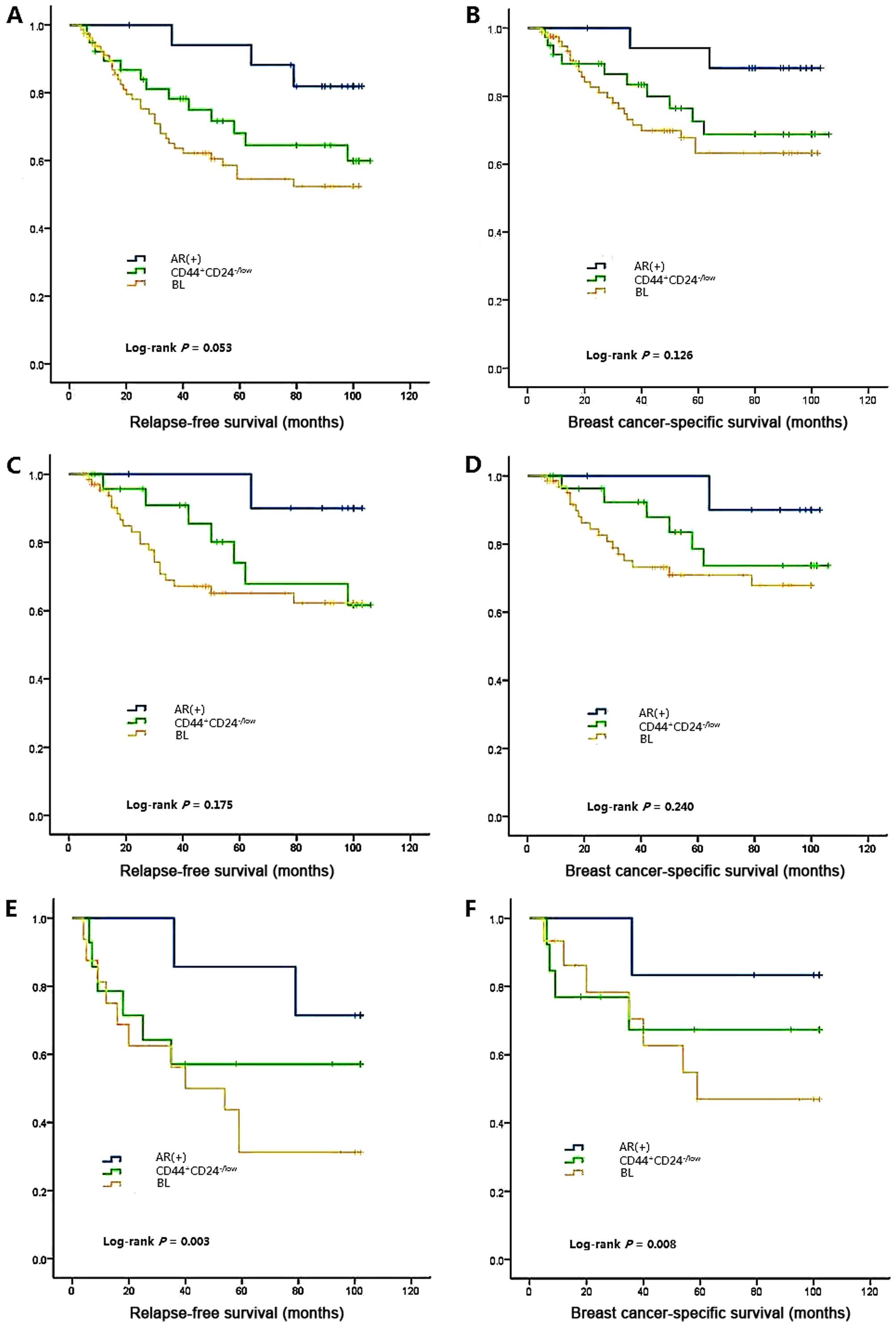
RFS and BCSS by IHC subtypes
The RFS time of TNBC patients ranged from 4 to 102
months with a median time of 61 months. During the study period, 50
out of 154 (32.5%) TNBC patients experienced local recurrence
and/or metastasis. Among these 50 cases, 32 (64%) were in the BL
group, 3 (6%) were in the AR+ group, 13 (26%) were in the
CD44+CD24-/low subtype, and 2 (4%) were in the
unclassified group. The hazard ratio (HR) and 95% confidence
interval (CI) of RFS for several basic characteristics by TNBC
subtype are shown in Table III.
Survival analyses are demonstrated in Fig. 3A. Larger tumor size, positive lymph
node status and higher histological grade significantly increased
the recurrence risk of TNBC tumors. All of the three subgroups
maintained this feature of TNBCs. However, marked tumor necrosis,
which could increase the recurrence risk of TNBC, AR+ and
CD44+CD24-/low subgroups, did not significantly affect the RFS
within the BL subgroup. A higher Ki67 labeling index (>30%) only
increased the recurrence risk of AR+ tumors.
 | Table III.Hazard ratios of triple-negative
breast cancer relapse-free survival for several basic
characteristics by immunohistochemistry subtypes. |
Table III.
Hazard ratios of triple-negative
breast cancer relapse-free survival for several basic
characteristics by immunohistochemistry subtypes.
|
|
|
| TNBC subtypes |
|---|
|
|
|
|
|
|---|
|
| TNBC n=154 |
| Basal-like
n=154 |
| AR+n=154 |
|
CD44+CD24−/low
n=154 |
|
|---|
|
|
|
|
|
|
|
|
|
|
|---|
| Variables | HR (95% CI) | P-value | HR (95% CI) | P-value | HR (95% CI) | P-value | HR (95% CI) | P-value |
|---|
| Age |
| 1.000 |
| 1.000 |
| 1.000 |
| 1.000 |
|
≤50 | 1.00 |
| 1.00 |
| 1.00 |
| 1.00 |
|
|
>50 | 0.88
(0.51–1.52) |
| 0.72
(0.48–1.39) |
| 1.06
(0.78–1.43) |
| 0.92
(0.63–1.5) |
|
| Pathological tumor
size |
| 0.012 |
| 0.032 |
| 0.002 |
| 0.019 |
|
pT1 | 1.00 |
| 1.00 |
| 1.00 |
| 1.00 |
|
|
pT2-3 | 2.63
(1.54–3.92) |
| 2.26
(1.32–3.49) |
| 3.08
(1.60–4.93) |
| 2.52
(1.22–3.85) |
|
| Lymph node
status |
| 0.003 |
| 0.024 |
| 0.001 |
| 0.014 |
|
Negative | 1.00 |
| 1.00 |
| 1.00 |
| 1.00 |
|
|
Positive | 2.86
(1.62–4.54) |
| 2.41
(1.32–3.54) |
| 3.13
(1.87–4.98) |
| 2.60
(1.43–3.87) |
|
| Histological
grade |
| 1.000 |
| 1.000 |
| 0.048 |
| 1.000 |
| 1 | 1.00 |
| 1.00 |
| 1.00 |
| 1.00 |
|
| 2 | 1.06
(0.53–2.24) |
| 1.02
(0.43–2.65) |
| 1.26
(1.09–2.87) |
| 1.03
(0.83–1.86) |
|
| 3 | 4,89
(2.37–8.65) | <0.001 | 3.58
(2.79–6.94) | <0.001 | 5.19
(2.46–9.12) | <0.001 | 4.12
(2.03–7.45) | <0.001 |
| Necrosis |
| 0.034 |
| 0.193 |
| 0.004 |
| 0.028 |
| Minimal
or absent | 1.00 |
| 1.00 |
| 1.00 |
| 1.00 |
|
|
Marked | 2.45
(1.14–3.96) |
| 1.52
(1.06–3.34) |
| 3.06
(1.65–4.12) |
| 2.52
(1.21–3.84) |
|
| Ki67 |
| 1.000 |
| 1.000 |
| 0.046 |
| 1.000 |
|
≤30% | 1.00 |
| 1.00 |
| 1.00 |
| 1.00 |
|
|
>30% | 1.07
(0.56–1.74) |
| 0.88
(0.54–1.45) |
| 1.64
(1.08–2.90) |
| 1.03
(0.84–1.79) |
|
| Adjuvant
chemotherapy |
| 1.000 |
| 0.004 |
| 0.942 |
| 0.246 |
| No | 1.00 |
| 1.00 |
| 1.00 |
| 1.00 |
|
|
Yes | 0.61
(0.32–1.34) |
| 0.26
(0.12–0.71) |
| 0.89
(0.64–1.35) |
| 2.15
(0.73–6.32) |
|
The BCSS time ranged from 2 to 108 months with a
median time of 68 months. Thirty-six of the 154 (23.4%) TNBC
patients succumbed to breast cancer, 12 patients succumbed to other
diseases and 106 were alive at the end of the study. The HR and 95%
CI for BCSS are shown in Table IV,
and survival analyses are shown in Fig.
3B. The three subtypes did not exhibit notable differences
either in the RFS or BCSS time (log-rank P=0.053 for RFS, log-rank
P=0.126 for BCSS). Multiple comparison only detected a difference
between the AR+ and BL group (log-rank P=0.020 for RFS, log-rank
P=0.044 for BCSS). Tumor size, lymph node involvement, histological
grade and tumor necrosis were significant prognostic factors in the
analysis with all cases of TNBC, and with each subtype of TNBC. In
the AR+ group, a higher Ki67 labeling index (>30%) also
demonstrated prognostic value.
 | Table IV.Hazard ratios of breast
cancer-specific survival in triple-negative breast cancer for
several basic characteristics by immunohistochemistry subtypes. |
Table IV.
Hazard ratios of breast
cancer-specific survival in triple-negative breast cancer for
several basic characteristics by immunohistochemistry subtypes.
|
| TNBC subtypes |
|---|
|
|
|
|---|
|
| TNBC n=154 |
| Basal-like
n=83 |
| AR+n=18 |
|
CD44+CD24−/low
n=39 |
|
|---|
| Variables | HR (95% CI) | P-value | HR (95% CI) | P-value | HR (95% CI) | P-value | HR (95% CI) | P-value |
|---|
| Age |
|
1.000 |
|
1.000 |
|
1.000 |
|
1.000 |
|
≤50 | 1.00 |
| 1.00 |
| 1.00 |
| 1.00 |
|
|
>50 | 1.06
(0.52–1.62) |
| 0.81
(0.38–1.54) |
| 1.10
(0.86–1.52) |
| 0.96
(0.58–1.46) |
|
| Pathological |
| <0.001 |
|
0.016 |
| <0.001 |
|
0.010 |
|
pT1 | 1.00 |
| 1.00 |
| 1.00 |
| 1.00 |
|
|
pT2-3 | 2.87
(1.64–4.26) |
| 2.44
(1.54–3.98) |
| 3.46
(1.87–5.65) |
| 2.61
(1.31–3.92) |
|
| Lymph node
status |
| <0.001 |
|
0.012 |
| <0.001 |
| <0.001 |
|
Negative | 1.00 |
| 1.00 |
| 1.00 |
| 1.00 |
|
Positive | 3.14
(1.81–5.96) |
| 2.53
(1.22–4.66) |
| 3.52
(1.84–5.26) |
| 2.84
(1.38–4.89) |
|
| Histological
grade |
|
1.000 |
|
0.076 |
|
|
|
1.000 |
| 1 | 1.00 |
| 1.00 |
| 1.00 |
| 1.00 |
|
| 2 | 1.12
(0.63–2.57) |
| 1.36
(0.87–2.84) |
| 1.32
(1.13–3.37) |
0.039 | 1.05
(0.75–1.97) |
|
| 3 | 5.23
(2.74–9.16) | <0.001 | 3.72
(2.97–7.42) | <0.001 | 6.21
(2.06–10.26) | <0.001 | 4.34
(2.42–8.16) | <0.001 |
| Necrosis |
|
0.019 |
|
0.042 |
| <0.001 |
|
0.008 |
| Minimal
or absent | 1.00 |
| 1.00 |
| 1.00 |
| 1.00 |
|
|
Marked | 2.53
(1.10–4.13) |
| 1.93
(1.33–4.16) |
| 3.54
(1.97–5.40) |
| 2.83
(1.46–3.99) |
|
| Ki67 |
|
1.000 |
|
1.000 |
|
0.032 |
|
1.000 |
|
≤30% | 1.00 |
| 1.00 |
| 1.00 |
| 1.00 |
|
|
>30% | 1.17
(0.46–1.95) |
| 0.92
(0.67–1.76) |
| 1.76
(1.12–3,23) |
| 1.13
(0.82–1.96) |
|
| Adjuvant
chemotherapy |
|
0.084 |
| <0.001 |
|
0.803 |
|
0.486 |
|
Not | 1.00 |
| 1.00 |
| 1.00 |
| 1.00 |
|
|
Yes | 0.52
(0.21–0.94) |
| 0.18
(0.09–0.65) |
| 0.92
(0.58–1.42) |
| 1.87
(0.96–4.86) |
|
Chemotherapy effects on the
subtypes
The univariate analyses above tested the prognostic
value of adjuvant chemotherapy in all TNBC cases and the different
subcategories, and it was revealed that only the BL group received
a significant RFS and BCSS benefit from adjuvant chemotherapy (RFS:
HR, 0.26; 95% CI, 0.12–0.71; P=0.004; BCSS: HR, 0.18; 95% CI,
0.09–0.65; P<0.001), whereas adjuvant chemotherapy was not
associated with significantly prolonged RFS and BCSS in other
subtypes and TNBCs as a whole. In order to investigate whether the
three subtypes responded differently to chemotherapy, we further
divided each subtype into two groups in the survival analysis
depending on use of adjuvant chemotherapy. Among the 83 BL
patients, 31 were treated with anthracycline-based chemotherapy (19
with doxorubicin/cyclophophamide and 12 with
fluorouracil/doxorubicin/cyclophosphamide), 38 were treated with
nonanthracycline-based chemotherapy (cyclophosphamide, methotrexate
and fluorouracil), and 14 received no adjuvant systemic therapy.
Among the 18 AR+ patients, 9 received anthracycline-based
chemotherapy (3 doxorubicin/cyclophosphamide and 6
fluorouracil/doxorubicin/cyclophosphamide), 5 received
nonanthracycline-based chemotherapy, and 4 received no adjuvant
systemic therapy. Among the 39 CD44+CD24-/low patients,
21 received anthracycline-based chemotherapy (14
doxorubicin/cyclophosphamide and 7
fluorouracil/doxorubicin/cyclophosphamide), 8 received
nonanthracycline-based chemotherapy, and 10 received no adjuvant
systemic therapy.
The survival analysis revealed that patients in the
BL group without chemotherapy had the shortest RFS and BCSS times
and demonstrated a significant survival gain following chemotherapy
(P=0.003 for RFS, P<0.001 for BCSS; Fig. 3C-F). Conversely, AR+ and
CD44+CD24-/low patients did not demonstrate a chemotherapy benefit
in either RFS or BCSS. However, the results require careful
interpretation due to the small numbers. There was no difference in
RFS and BCSS among the three subclasses; however, after we
categorized each subclass according to chemotherapy, a notable
distinction emerged (log-rank P=0.003 for RFS, log-rank P=0.008 for
BCSS).
In the multiple variate analyses (adjusted for age,
tumor size, histological grade, lymph node status and tumor
necrosis), the BL group demonstrated a significantly poorer
survival, with a HR of 2.98 vs. the luminal A cohort (95% CI,
1.38–6.10; P<0.001; Table VA), a
higher HR of 3.81 vs. luminal A in the cases without chemotherapy
(95% CI, 1.98–6.32; P<0.001; Table
VC), and a relatively lower HR of 1.93 vs. luminal A in the
cases with chemotherapy (95% CI, 1.21–4.09, P=0.028; Table VB). This confirmed the survival gain
of adjuvant chemotherapy in BL patients. In contrast, the AR+ group
did not exhibit a poorer survival vs. the luminal A cohort (HR,
1.38; 95% CI, 0.67–2.14; P=0.204), and the HR was 1.13 (95% CI,
0.62–2.89; P=0.574) in the cases with chemotherapy, and 1.53 (95%
CI, 0.68–1.98; P=0.124) in the cases without chemotherapy. The
decrease in HR owing to chemotherapy in the AR+ group (from 1.53 to
1.13) was far less significant than that in the BL group (from 3.81
to 1.93). In the CD44+CD24-/low group, there was an
increase rather than a decrease in HR in the subset of patients who
received chemotherapy (HR, 2.30; 95% CI, 0.95–2.84; P=0.003)
compared with those who did not (HR, 1.72; 95% CI, 0.88–2.74;
P=0.092). We cannot assume that chemotherapy increases the risk of
mortality in CD44+CD24-/low phenotype TNBCs, but the results did
reveal a notable trait of CD44+CD24-/low tumors in that
they do not respond to chemotherapy as well as the BL subtype.
 | Table V.Cox regression analysis to estimate
adjusted hazard ratios of breast cancer subtypes. |
Table V.
Cox regression analysis to estimate
adjusted hazard ratios of breast cancer subtypes.
| A, Cox regression
analysis of all 1646 cases |
|---|
|
|---|
|
| Relapse-free
survival | Breast
cancer-specific survival |
|---|
|
|
|
|
|---|
| Subtypes | HR (95% CI) | P-value | HR (95% CI) | P-value |
|---|
| IHC-Luminal A | 1.00 |
| 1.00 |
|
| IHC-HER2 | 2.96
(1.23–4.87) | <0.001 | 3.13
(2.28–5.07) | <0.001 |
| IHC-TNBC | 2.04
(1.11–4.38) | 0.017 | 2.15
(1.43–4.16) | 0.008 |
|
IHC-TN/BL | 2.85
(1.18–5.02) | <0.001 | 2.98
(1.38–6.10) | <0.001 |
|
IHC-TN/AR+ | 1.12
(0.87–1.68) | 0.541 | 1.38
(0.67–2.14) | 0.204 |
|
IHC-TN/CD44+CD24−/low | 1.78
(1.07–3.20) | 0.073 | 1.86
(1.18–3.75) | 0.052 |
|
IHC-TN/unassigned | 1.26
(1.11–2.45) | 0.296 | 1.43
(1.06–2.82) | 0.187 |
|
| B, Cox regression
analysis of 1360 cases treated with adjuvant chemotherapy. |
|
|
| Relapse-free
survival | Breast
cancer-specific survival |
|
|
|
|
| Subtypes | HR (95% CI) | P-value | HR (95% CI) | P-value |
|
| IHC-Luminal A | 1.00 |
| 1.00 |
|
| IHC-HER2 | 2.61
(1.24–4.78) | <0.001 | 2.72
(1.52–5.12) | <0.001 |
| IHC-TNBC | 1.93
(1.13–3.34) | 0.031 | 2.11
(1.28–3.84) | 0.019 |
|
IHC-TN/BL | 1.89
(1.17–3.96) | 0.048 | 1.93
(1.21–4.09) | 0.028 |
|
IHC-TN/AR+ | 1.09
(0.45–1.56) | 0.622 | 1.13
(0.62–2.89) | 0.574 |
|
IHC-TN/CD44+CD24−/low | 2.17
(0.76–2.74) | 0.006 | 2.30
(0.95–2.84) | 0.003 |
|
IHC-TN/unassigned | 1.14
(0.45–2.35) | 0.507 | 1.31
(0.61–2.72) | 0.232 |
|
| C, Cox regression
analysis of 286 cases treated without chemotherapy. |
|
|
| Relapse-free
survival | Breast
cancer-specific survival |
|
|
|
|
| Subtypes | HR (95% CI) | P-value | HR (95% CI) | P-value |
|
| IHC-Luminal A | 1.00 |
| 1.00 |
|
| IHC-HER2 | 3.32
(1.32–5.14) | <0.001 | 3.48
(1.15–5.54) | <0.001 |
| IHC-TNBC | 2.19
(1.11–4.98) | 0.002 | 2.25
(1.08–5.11) | <0.001 |
|
IHC-TN/BL | 3.64
(1.86–7.65) | <0.001 | 3.81
(1.98–6.32) | <0.001 |
|
IHC-TN/AR+ | 1.25
(0.76–2.24) | 0.287 | 1.53
(0.68–1.98) | 0.124 |
|
IHC-TN/CD44+CD24−/low | 1.68
(1.14–3.07) | 0.115 | 1.72
(0.88–2.74) | 0.092 |
|
IHC-TN/unassigned | 1.37
(0.64–2.73) | 0.196 | 1.48
(0.49–2.64) | 0.163 |
Correlation between IHC TNBC subtypes
and subtypes in non-TNBC
CK5/6+, CK14+ and CD44+CD24-/low
phenotype were not only observed in TNBCs, but also in non-TNBC
cases. Of the 1492 non-TNBCs, 34 cases positively expressed either
CK5/6 or CK14, and they are referred to as BL/non-TN in this study.
Accordingly, the 36 cases that had the CD44+CD24-/low
phenotype are referred to as CD44+CD24-/low/non-TN. An
issue that cannot be ignored is the correlation between BL tumors
that are TNBC (BL/TN) and BL/non-TN, and CD44+CD24-/low
tumors that are TNBC (CD44+CD24-/low/TN) and
CD44+CD24-/low/non-TN. To be specific, we take the BL
subtype as an example. BL was defined as positive for CK5/6 or
CK14, and BL/TN has certain distinct features as shown above,
including younger age, higher histological grade and poorer
prognosis. Therefore, through a comparison of clinicopathological
characteristics between BL/TN and BL/non-TN we could observe
whether these features of CK5/6+ and/or CK14+ tumors would be
retained regardless of their clinical ER, PR and HER2 status, and
particularly their TN status. The results of comparison may
indicate whether BL/TN cases possess these traits more due to their
BL status (i.e. their positivity for CK5/6 or CK14) or more due to
their TN status (i.e. their triple-negative status). Thus, it may
provide us with a better understanding of the intrinsic quality of
these TNBC subtypes. A comparison of clinicopathological
characteristics between BL/TN and BL/non-TN, and
CD44+CD24-/low/TN and CD44+CD24-/low/non-TN
is shown in Table V. As for the AR+
group, we did not compare AR+ tumors that were triple-negative with
those that were not. As mentioned in the Patients and methods
section, several previous studies (10,13,20,21)
have made the same observation that TN tumors with high AR protein
and/or gene expression (or LAR, according to Lehmann et al
(10,12) were usually identified as HER2 or
luminal by PAM50 intrinsic subtyping, and their levels of AR
expression resembled the levels observed in HER2 and ER-positive
tumors that were not TN. However, the authors had divergent
opinions on percentage that the HER2 and luminal groups accounted
for. According to Mayer et al (19), the LAR subtype is classified as HER2
(74.3%) and luminal (14.3%); however, based on the statistics of
Lehmann et al (10) 82% of LAR
cases were luminal (either luminal A or B). Therefore, we
separately compared the AR+ group of TNBC with the luminal subtype
[including luminal A (ER+ and/or PR+,
HER2−) and luminal B (ER+ and/or
PR+, HER2+)] and HER2 subtype
(ER−, PR−, HER2+) that were not
TNBC. Based on the data from Table
VI, BL/TN cases demonstrated almost undistinguishable
clinicopathological characteristics compared with BL/non-TN cases,
as did CD44+CD24-/low/TN cases compared with
CD44+CD24-/low/non-TN cases. The features of the AR+
group resembled those of the non-TNBC luminal group rather than
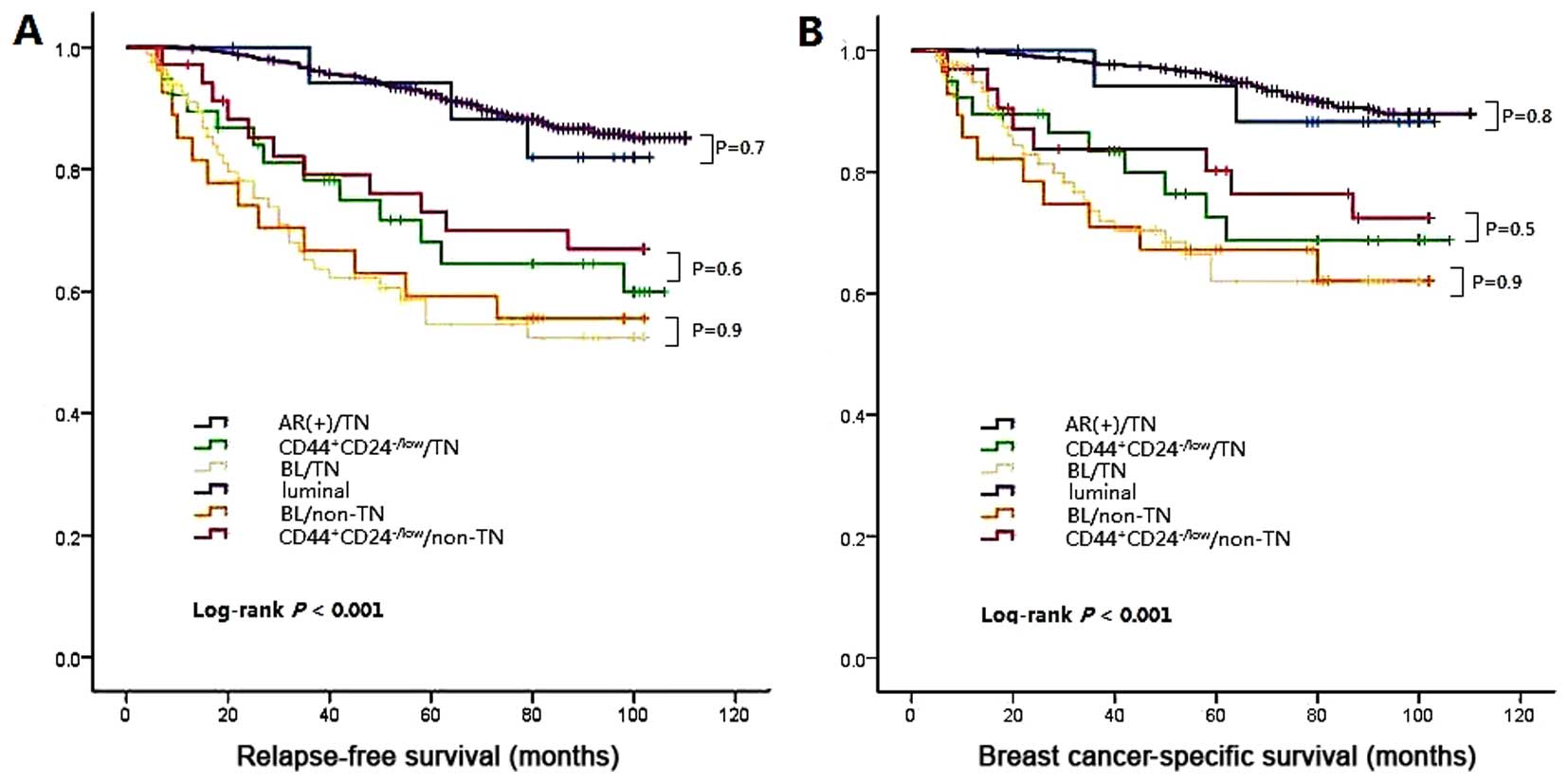
those of HER2. Next, a survival analysis was performed and
differences in RFS and BCSS were compared between BL/TN and
BL/non-TN, CD44+CD24-/low/TN and
CD44+CD24-/low/non-TN, and the AR+ and luminal group
(Fig. 4). Multiple comparison
revealed no significant difference between BL/TN and BL/non-TN
(log-rank P=0.9 for RFS, log-rank P=0.9 for BCSS),
CD44+CD24-/low/TN and CD44+CD24-/low/non-TN
(log-rank P=0.6 for RFS, log-rank P=0.5 for BCSS), or the AR+ group
and luminal group (log-rank P=0.7 for RFS, log-rank P=0.8 for
BCSS).
 | Table VI.Correlation between the
triple-negative and non-triple negative breast cancer
immunohistochemistry subtypes. |
Table VI.
Correlation between the
triple-negative and non-triple negative breast cancer
immunohistochemistry subtypes.
|
| BL/TN | BL/TN |
| CD44+
CD24−/low/TN | CD44+
CD24−/low/non-TN |
|
AR+/TN | Luminal |
| AR+/TN | HER2 |
|
|---|
|
|
|
|
|
|
|
|
|
|
|---|
| Variables | n=83 | n=34 | P-value | n=39 | n=36 | P-value | n=18 | n=965 | P-value | n=18 | n=344 | P-value |
|---|
| Age |
|
| 0.834 |
|
| 0.926 |
|
| 0.818 |
|
| 0.073 |
|
≤50 | 52 | 22 |
| 18 | 17 |
| 6 | 347 |
| 6 | 189 |
|
|
>50 | 31 | 12 |
| 21 | 19 |
| 12 | 618 |
| 12 | 155 |
|
| Family history |
|
| 0.575 |
|
| 0.775 |
|
|
|
|
| 0.924 |
| No | 76 | 30 |
| 35 | 33 |
| 17 | 896 |
| 17 | 323 |
|
|
Yes | 7 | 4 |
| 4 | 3 |
| 1 | 69 |
| 1 | 21 |
|
| Histological
type |
|
|
|
|
|
|
|
|
|
|
|
|
|
Invasive ductal carcinoma | 63 | 21 |
| 14 | 15 |
| 6 | 872 |
| 6 | 319 |
|
|
Invasive lobular
carcinoma | 2 | 3 |
| 1 | 2 |
| 1 | 49 |
| 1 | 11 |
|
|
Medullary carcinoma | 11 | 6 |
| 2 | 0 |
| 0 | 12 |
| 0 | 7 |
|
|
Metaplastic carcinoma | 3 | 1 |
| 19 | 18 |
| 0 | 10 |
| 0 | 0 |
|
|
Apocrine carcinoma | 2 | 0 |
| 0 | 0 |
| 11 | 14 |
| 11 | 0 |
|
|
Others | 2 | 3 |
| 3 | 1 |
| 0 | 8 |
| 0 | 7 |
|
| Pathological tumor
size |
|
| 0.790 |
|
| 0.378 |
|
|
|
|
| 0.031 |
|
pT1 | 24 | 9 |
| 18 | 13 |
| 10 | 480 |
| 10 | 107 |
|
|
pT2-3 | 59 | 25 |
| 21 | 23 |
| 8 | 485 |
| 8 | 237 |
|
| Histological
grade |
|
| 0.764 |
|
| 0.836 |
|
| 0.561 |
|
| 0.019 |
| 1 | 3 | 2 |
| 7 | 5 |
| 9 | 363 |
| 9 | 74 |
|
| 2 | 12 | 6 |
| 11 | 12 |
| 6 | 396 |
| 6 | 166 |
|
| 3 | 68 | 26 |
| 21 | 19 |
| 3 | 206 |
| 3 | 104 |
|
| Lymph node
status |
|
| 0.609 |
|
| 0.345 |
|
| 0.710 |
|
| 0.318 |
|
Negative | 47 | 21 |
| 23 | 25 |
| 12 | 602 |
| 12 | 188 |
|
|
Positive | 36 | 13 |
| 16 | 11 |
| 6 | 363 |
| 6 | 156 |
|
| Lymphovascular
invasion |
|
| 0.787 |
|
| 0.798 |
|
| 0.964 |
|
|
0.808 |
|
Absent | 63 | 25 |
| 26 | 21 |
| 16 | 861 |
| 16 | 299 |
|
|
Present | 20 | 9 |
| 13 | 15 |
| 2 | 104 |
| 2 | 45 |
|
| Necrosis |
|
| 0.553 |
|
| 0.680 |
|
| 0.424 |
|
|
0.145 |
| Minimal
or absent | 24 | 8 |
| 22 | 22 |
| 15 | 725 |
| 15 | 230 |
|
|
Marked | 59 | 26 |
| 17 | 14 |
| 3 | 240 |
| 3 | 114 |
|
| Ki67 |
|
| 0.374 |
|
| 0.711 |
|
| 0.489 |
|
|
0.004 |
|
≤30% | 29 | 9 |
| 19 | 16 |
| 14 | 678 |
| 14 | 149 |
|
|
>30% | 54 | 25 |
| 20 | 20 |
| 4 | 287 |
| 4 | 195 |
|
| EGFR |
|
| 0.728 |
|
| 0.701 |
|
| 0.701 |
|
| <0.001 |
|
Negative | 41 | 18 |
| 30 | 29 |
| 8 | 473 |
| 8 | 281 |
|
|
Positive | 42 | 16 |
| 9 | 7 |
| 10 | 492 |
| 10 | 63 |
|
| E-cadherin |
|
| 0.321 |
|
| 0.192 |
|
| 0.621 |
|
|
0.499 |
|
Negative | 38 | 19 |
| 37 | 31 |
| 4 | 265 |
| 4 | 102 |
|
|
Positive | 45 | 15 |
| 2 | 5 |
| 14 | 700 |
| 14 | 242 |
|
| Vimentin |
|
| 0.650 |
|
| 0.261 |
|
| 0.596 |
|
|
0.747 |
|
Negative | 55 | 24 |
| 4 | 7 |
| 15 | 754 |
| 15 | 296 |
|
|
Positive | 28 | 10 |
| 35 | 29 |
| 3 | 211 |
| 3 | 48 |
|
| Claudin 3 |
|
| 0.118 |
|
| 0.611 |
|
| 0.906 |
|
|
0.898 |
|
Negative | 10 | 8 |
| 36 | 32 |
| 2 | 116 |
| 2 | 35 |
|
|
Positive | 73 | 26 |
| 3 | 4 |
| 16 | 849 |
| 16 | 309 |
|
| Claudin 4 |
|
| 0.318 |
|
| 0.834 |
|
| 0.647 |
|
|
0.646 |
|
Negative | 9 | 6 |
| 29 | 26 |
| 1 | 83 |
| 1 | 12 |
|
|
Positive | 74 | 28 |
| 10 | 10 |
| 17 | 882 |
| 17 | 332 |
|
| Claudin 7 |
|
| 0.827 |
|
| 0.233 |
|
| 0.771 |
|
|
0.458 |
|
Negative | 11 | 4 |
| 28 | 30 |
| 1 | 71 |
| 1 | 9 |
|
|
Positive | 72 | 30 |
| 11 | 6 |
| 17 | 894 |
| 17 | 335 |
|
| RFS event |
|
| 0.928 |
|
| 0.602 |
|
| 0.714 |
|
| 0.151 |
| No | 51 | 22 |
| 26 | 25 |
| 15 | 857 |
| 15 | 231 |
|
|
Yes | 32 | 12 |
| 13 | 11 |
| 3 | 108 |
| 3 | 113 |
|
| Mean survival
time | 75.8 | 77.7 |
| 84.7 | 86.1 |
| 96.3 | 103.2 |
| 96.3 | 79.3 |
|
| 95% CI | 59.9–88.4 | 62.8–91.4 |
| 73.4–94.2 | 75.6–96.6 |
| 84.0–105.7 | 93.3–109.2 |
| 84.0–105.7 | 60.5–93.6 |
Discussion
In this study, a large number of clinical breast
cancer cases were evaluated and the following observations
concerning TN breast cancers were made: i) TN disease is a
heterogeneous clinical entity composed of three main IHC subtypes,
with the BL tumor type predominating (>50%); ii) The three
subcategories demonstrated a statistically significant difference
with regard to age, tumor size, histological grade, tumor necrosis,
Ki67 labeling index and response to chemotherapy; iii) Basal-like
tumors that are TN exhibit almost undistinguishable
clinicopathological characteristics compared with BL tumors that
are non-TN. The same applies with
CD44+CD24-/low/TN vs.
CD44+CD24-/low/non-TN and
AR+/TN vs. luminal/non-TN.
Our study is a preliminary attempt to use gene
expression subtypes in a practical and clinically accessible
diagnostic test. We use IHC methodology to observe how TNBC can be
broken down into components. This novel IHC classification system
was based on the perspectives of Lehmann et al (10,12,19), who
identified six subtypes (BL1, BL2, IM, M, MSL and LAR), and Prat
et al (13), who contended
that the three main subtypes were BL, claudin-low and
luminal/HER2-enriched. These two seemingly different
classifications are correlated; for instance, LAR shares a number
of gene expression features of luminal and HER2-enriched cancers,
as illustrated in Patients and methods. However, in the definition
of Prat et al (13), the
identification of luminal/TN tumors, HER2/TN tumors might appear at
first glance to be counterintuitive, and an explanation is required
with regard to the discrepancy between gene expression and
IHC-based assays. One possibility is the false positivity or false
negativity of the IHC-based assays in determining hormone receptor
or HER2 status (28). Another
possibility is that the pathology and gene expression data could
have been obtained from two different areas of the same tumor
(i.e., intratumor heterogeneity) (29). The most plausible explanation is that
gene expression measures a large number of related genes, compared
with the three individual pathology-based biomarkers that define TN
disease (13). Thus, multigene
expression data using tens to hundreds of genes might better
capture the true biological profile of a given tumor compared with
three or four individual biomarkers (30). For example, a TN tumor that has low
levels of ESR1 and PGR, and consequently is ER- and PR- by IHC,
might be identified as luminal due to the high expression of other
luminal-related genes (i.e., AR, GATA3 and/or FOXA1) and the low
expression of basal- and proliferation-related genes. Another
example comes from the identification of HER2-enriched/TN tumors
that do not amplify ERBB2, some of which might be driven by high
EGFR (13).
In previous studies, BL breast cancers accounted for
up to 15% of all breast cancers (31,32). Most
of them used the definition of Nielsen et al (31), which is positive staining for CK5/6 or
EGFR (31). In this study, the
proportion of BL breast cancers was 7.11% (53.9% of TNBCs). Of a
total of 117 BL breast cancers, 70.94% (83 of 117) were TNBCs, and
29.06% (34 of 117) were non-TNBCs. We used basal markers CK5/6 and
CK14 instead of CK5/6 and EGFR for four reasons: i) Based on the
recent progress in TNBC gene subtyping by Prat et al
(13), expression of EGFR was
observed to be significantly increased in HER2-enriched/TN tumors
compared with HER2-enriched/non-TN tumors, thus suggesting that
certain HER2-enriched tumors, which are at gene level in line with
HER2-enriched tumors but are HER2- by IHC, may be driven by EGFR as
discussed above. This implies that EGFR expression is not confined
to BL cancers (10,19). ii) The EGFR gene is not enriched in
all BL tumors but in the BL2 subtype alone (10). It is also enriched in a minority of
mesenchymal subtypes (10). iii) Not
only has specificity of EGFR for defining BL breast cancers become
lower than it used to be when subtyping was not as comprehensive as
today, but also the prognostic value of EGFR was challenged. In the
study of Choi et al (33),
CK5/6 was a poor prognostic marker whereas EGFR was not. 4)
According to Won et al (34),
in a survey of IHC biomarkers for BL breast cancer against a gene
expression profile gold standard, CK14 was the most specific
(specificity 100%) among the 46 biomarkers surveyed. If we used
CK5/6 and EGFR, the proportion of BL breast cancers increased to
14.12%, and accounted for 65.6% of TNBCs, which was similar to the
figure of 15% obtained in the previous study. However, this might
obscure certain significant information since EGFR+ tumors comprise
part of the AR+ group. Indeed, 10 out of 18 AR+ TNBCs demonstrated
weak or strong positivity for EGFR.
AR+ tumors constitute a distinct subgroup of TNBC. A
total of 11.7% of TNBCs were AR+ in this study. Among 22 studies
summarized in the article of Safarpour et al (35), the proportion of tumors with positive
AR among TNBCs ranged from 6.6% to 75%. Among six studies which had
used the most recent ASCO/CAP guidelines (1% and more) for ER, PR
and AR positivity, the expression rate of AR ranged from 12.7% to
41.4%. This group has certain valuable clinicopathological features
including smaller tumor size, higher median age, lower histological
grade, higher percentage of apocrine morphology, lower
proliferation index (measured by Ki67) and statistically longer
disease-free survival and overall survival (8,36–48). Our study also arrived at similar
conclusions. In terms of IHC features, it is noteworthy that none
of the AR+ tumors in our study were positive for CK5/6 or CK14, and
due to the relatively small series of analyzed samples this may be
coincidental; however, it is in accordance with the study of
Lehmann et al that LAR cancers lacked expression of basal
cytokeratins. So far, no organization has recommended AR assessment
for breast cancers; however, we support routine assessment of AR at
least for TNBCs considering the predictive value of AR in TNBC.
AR+ and BL are two subtypes that have received
significant interest and are relatively well analyzed. However,
emerging data imply that TN disease is a broad and diverse category
for which additional subclassifications are required. One of the
contributions of this study is that, for the first time, we
distinguished a subgroup of TNBC as the CD44+CD24-/low
phenotype using IHC markers, and the overlap between this third
group and BL and AR+ was low (3 and 0 cases, respectively).
CD44+CD24-/low is a marker of breast stem cells and
tumor-initiating cells and is observed to be exclusively enriched
in claudin-low subtype (26,27). There are also other features in the
claudin-low subtype, for instance, low gene expression of tight
junction proteins claudin 3, 4 and 7 and E-cadherin (9,26,27,49,50).
However, when we used negativity for claudins 3, 4 and 7 to define
the third group, there were 24 cases, a relatively large
proportion, that could not be classified. This is possibly due to
the fact that negativity for all claudins is a much stricter
restriction compared with CD44+CD24-/low. In addition, a
study of Prat et al (9), a
researcher who contributed significantly to our knowledge of the
claudin-low subtype of breast cancer, revealed that BL tumors did
not demonstrate significantly lower expression of CD24 as a group.
This crucial distinction may explain the lowest overlap between the
BL group and CD44+CD24-/low group. In another
classification where vimentin+ and E-cadherin- were used, the
highest overlap was observed. In fact, undifferentiated levels of
mesenchymal (vimentin) markers exist not only within the
claudin-low subtype, but also in BL breast cancers, and no
statistically significant difference was observed between
claudin-low and BL tumors (9). We
defined a number of differences between the
CD44+CD24-/low subtype and the other groups.
Clinicopathological characteristics including histological grade
and tumor necrosis were different from the AR+ as well as the BL
group. The age at diagnosis of this group was older, the tumor size
was smaller, and the Ki67 labeling index was lower than that of the
BL group. The two groups demonstrated an unfavorable clinical
outcome; however, the CD44+CD24-/low group did not
benefit from adjuvant chemotherapy to the extent that the BL group
did. Sabatier et al (51) also
made similar findings in their study of clinical, pathological and
prognostic characterization of claudin-low breast cancers,
revealing that the percentage of patients older than 50, the
percentage of grade 3 claudin-low tumors, the percentage of tumors
measuring 2 cm or less, and the 5-year disease-free survival rate
were all intermediate between that of the highly proliferative
subtypes (BL and HER2-enriched) and that of less proliferative ones
(luminal A and normal).
Without chemotherapy, the BL subcategory had the
poorest prognosis in terms of RFS and BCSS. Notably, the BL group
demonstrated a distinct clinical benefit with standard adjuvant
chemotherapy. Conversely, adjuvant chemotherapy demonstrated little
clinical benefit for the AR+ and CD44+CD24-/low
subclasses. Masuda et al (52)
performed a retrospective analysis on 130 TNBC cases treated with
neoadjuvant adriamycin/cytoxan/taxol-containing chemotherapy, and
subtype-specific responses differed substantially, with the BL1
subtype achieving the highest pathological complete remission rate
(52%), and the BL2, LAR and MSL subtypes having the lowest
responses (0%, 10% and 23%, respectively). In accordance with the
work of Masuda et al (52),
Mayer et al (19) observed a
similar distribution of subtype-specific differences in survival.
These findings should guide differential use of chemotherapy-based
regimens and instruct clinical trials to investigate targeted
therapies.
In summary, TNBC is a relatively uncommon, notably
aggressive disease, and there is a major requirement to better
decipher the heterogeneity of TNBC in order to tackle the
challenges in combatting this disease. New therapeutic strategies
for TNBC are emerging since gene subtyping was identified.
Therefore, our future clinical trial design for TNBC intends to
focus on continued efforts to translate genetic approaches into
clinical utility, to develop a more standard IHC classification of
TNBC. Our aim is to provide a labor- and timesaving method for
clinicians to distinguish the subtypes of TNBC in their daily work
and, in the near future, select a more appropriate personalized
therapy based on these subtypes.
Acknowledgements
This study was supported by the National Natural
Science Foundation of China (grant no. 81472343).
References
|
1
|
Cheang MC, Voduc D, Bajdik C, Leung S,
McKinney S, Chia SK, Perou CM and Nielsen TO: Basal-like breast
cancer defined by five biomarkers has superior prognostic value
than triple-negative phenotype. Clin Cancer Res. 14:1368–1376.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Aguiar FN, Mendes HN, Cirqueira CS, Bacchi
CE and Carvalho FM: Basal cytokeratin as a potential marker of low
risk of invasion in ductal carcinoma in situ. Clinics (Sao Paulo).
68:638–643. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Fadare O, Wang SA and Hileeto D: The
expression of cytokeratin 5/6 in invasive lobular carcinoma of the
breast: evidence of a basal-like subset? Hum Pathol. 39:331–336.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lee HJ, Seo AN, Kim EJ, Jang MH, Kim YJ,
Kim JH, Kim SW, Ryu HS, Park IA, Im SA, et al: Prognostic and
predictive values of EGFR overexpression and EGFR copy number
alteration in HER2-positive breast cancer. Br J Cancer.
112:103–111. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Park HS, Jang MH, Kim EJ, Kim HJ, Lee HJ,
Kim YJ, Kim JH, Kang E, Kim SW, Kim IA and Park SY: High EGFR gene
copy number predicts poor outcome in triple-negative breast cancer.
Mod Pathol. 27:1212–1222. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Rakha EA, El-Sayed ME, Green AR, Lee AH,
Robertson JF and Ellis IO: Prognostic markers in triple-negative
breast cancer. Cancer. 109:25–32. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Sood N and Nigam JS: Correlation of CK5
and EGFR with clinicopathological profile of triple-negative breast
cancer. Patholog Res Int. 2014:1418642014.PubMed/NCBI
|
|
8
|
Niemeier LA, Dabbs DJ, Beriwal S, Striebel
JM and Bhargava R: Androgen receptor in breast cancer: expression
in estrogen receptor-positive tumors and in estrogen
receptor-negative tumors with apocrine differentiation. Mod Pathol.
23:205–212. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Prat A, Parker JS, Karginova O, Fan C,
Livasy C, Herschkowitz JI, He X and Perou CM: Phenotypic and
molecular characterization of the claudin-low intrinsic subtype of
breast cancer. Breast Cancer Res. 12:R682010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lehmann BD, Bauer JA, Chen X, Sanders ME,
Chakravarthy AB, Shyr Y and Pietenpol JA: Identification of human
triple-negative breast cancer subtypes and preclinical models for
selection of targeted therapies. J Clin Invest. 121:2750–2767.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Turner NC and Reis-Filho JS: Tackling the
diversity of triple-negative breast cancer. Clin Cancer Res.
19:6380–6388. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lehmann BD and Pietenpol JA:
Identification and use of biomarkers in treatment strategies for
triple-negative breast cancer subtypes. J Pathol. 232:142–150.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Prat A, Adamo B, Cheang MC, Anders CK,
Carey LA and Perou CM: Molecular characterization of basal-like and
non-basal-like triple-negative breast cancer. Oncologist.
18:123–133. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Abramson VG, Lehmann BD, Ballinger TJ and
Pietenpol JA: Subtyping of triple-negative breast cancer:
implications for therapy. Cancer. 121:8–16. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Schmadeka R, Harmon BE and Singh M:
Triple-negative breast carcinoma: current and emerging concepts. Am
J Clin Pathol. 141:462–477. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Choo JR and Nielsen TO: Biomarkers for
basal-like breast cancer. Cancers (Basel). 2:1040–1065. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ricardo S, Vieira AF, Gerhard R, Leitão D,
Pinto R, Cameselle-Teijeiro JF, Milanezi F, Schmitt F and Paredes
J: Breast cancer stem cell markers CD44, CD24 and ALDH1: expression
distribution within intrinsic molecular subtype. J Clin Pathol.
64:937–946. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Mylona E, Giannopoulou I, Fasomytakis E,
Nomikos A, Magkou C, Bakarakos P and Nakopoulou L: The
clinicopathologic and prognostic significance of
CD44+/CD24(−/low) and
CD44−/CD24+ tumor cells in invasive breast
carcinomas. Hum Pathol. 39:1096–1102. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Mayer IA, Abramson VG, Lehmann BD and
Pietenpol JA: New strategies for triple-negative breast cancer -
deciphering the heterogeneity. Clin Cancer Res. 20:782–790. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Neve RM, Chin K, Fridlyand J, Yeh J,
Baehner FL, Fevr T, Clark L, Bayani N, Coppe JP, Tong F, et al: A
collection of breast cancer cell lines for the study of
functionally distinct cancer subtypes. Cancer Cell. 10:515–527.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kao J, Salari K, Bocanegra M, Choi YL,
Girard L, Gandhi J, Kwei KA, Hernandez-Boussard T, Wang P, Gazdar
AF, et al: Molecular profiling of breast cancer cell lines defines
relevant tumor models and provides a resource for cancer gene
discovery. PLoS One. 4:e61462009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Liu T, Zhang X, Shang M, Zhang Y, Xia B,
Niu M, Liu Y and Pang D: Dysregulated expression of Slug, vimentin
and E-cadherin correlates with poor clinical outcome in patients
with basal-like breast cancer. J Surg Oncol. 107:188–194. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Lee J, Hahm ER, Marcus AI and Singh SV:
Withaferin A inhibits experimental epithelial-mesenchymal
transition in MCF-10A cells and suppresses vimentin protein level
in vivo in breast tumors. Mol Carcinog. 54:417–429. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Rito M, Schmitt F, Pinto AE and André S:
Fibromatosis-like metaplastic carcinoma of the breast has a
claudin-low immunohistochemical phenotype. Virchows Arch.
465:185–191. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhang Y, Toy KA and Kleer CG: Metaplastic
breast carcinomas are enriched in markers of tumor-initiating cells
and epithelial to mesenchymal transition. Mod Pathol. 25:178–184.
2012.PubMed/NCBI
|
|
26
|
Giatromanolaki A, Sivridis E, Fiska A and
Koukourakis MI: The CD44+/CD24- phenotype relates to
‘triple-negative’ state and unfavorable prognosis in breast cancer
patients. Med Oncol. 28:745–752. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Honeth G, Bendahl PO, Ringner M, Ringnér
M, Saal LH, Gruvberger-Saal SK, Lövgren K, Grabau D, Fernö M, Borg
A and Hegardt C: The CD44+/CD24-phenotype is enriched in basal-like
breast tumors. Breast Cancer Res. 10:R532008. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Hammond ME, Hayes DF, Wolff AC, Mangu PB
and Temin S: American Society of Clinical Oncology/College of
American Pathologists guideline recommendations for
immunohistochemical testing of estrogen and progesterone receptors
in breast cancer. J Oncol Pract. 6:195–197. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Barry WT, Kernagis DN, Dressman HK,
Griffis RJ, Hunter JD, Olson JA, Marks JR, Ginsburg GS, Marcom PK,
Nevins JR, et al: Intratumor heterogeneity and precision of
microarray-based predictors of breast cancer biology and clinical
outcome. J Clin Oncol. 28:2198–2206. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Prat A, Parker JS, Fan C and Perou CM:
PAM50 assay and the three-gene model for identifying the major and
clinically relevant molecular subtypes of breast cancer. Breast
Cancer Res Treat. 135:301–306. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Nielsen TO, Hsu FD, Jensen K, Cheang M,
Karaca G, Hu Z, Hernandez-Boussard T, Livasy C, Cowan D, Dressler
L, et al: Immunohistochemical and clinical characterization of the
basal-like subtype of invasive breast carcinoma. Clin Cancer Res.
10:5367–5374. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Badve S, Dabbs DJ, Schnitt SJ, Baehner FL,
Decker T, Eusebi V, Fox SB, Ichihara S, Jacquemier J, Lakhani SR,
et al: Basal-like and triple-negative breast cancers: a critical
review with an emphasis on the implications for pathologists and
oncologists. Mod Pathol. 24:157–167. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Choi YL, Oh E, Park S, Kim Y, Park YH,
Song K, Cho EY, Hong YC, Choi JS, Lee JE, et al: Triple-negative,
basal-like and quintuple-negative breast cancers: better prediction
model for survival. BMC Cancer. 10:5072010. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Won JR, Gao D, Chow C, Cheng J, Lau SY,
Ellis MJ, Perou CM, Bernard PS and Nielsen TO: A survey of
immunohistochemical biomarkers for basal-like breast cancer against
a gene expression profile gold standard. Mod Pathol. 26:1438–1450.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Safarpour D, Pakneshan S and Tavassoli FA:
Androgen receptor (AR) expression in 400 breast carcinomas: is
routine AR assessment justified? Am J Cancer Res. 4:353–368.
2014.PubMed/NCBI
|
|
36
|
McNamara KM, Yoda T, Miki Y, Chanplakorn
N, Wongwaisayawan S, Incharoen P, Kongdan Y, Wang L, Takagi K, Mayu
T, et al: Androgenic pathway in triple negative invasive ductal
tumors: its correlation with tumor cell proliferation. Cancer Sci.
104:639–646. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Tsutsumi Y: Apocrine carcinoma as
triple-negative breast cancer: novel definition of apocrine-type
carcinoma as estrogen/progesterone receptor-negative and androgen
receptor-positive invasive ductal carcinoma. Jpn J Clin Oncol.
42:375–386. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Thike AA, Yong-Zheng Chong L, Cheok PY, Li
HH, Wai-Cheong Yip G, Huat Bay B, Tse GM, Iqbal J and Tan PH: Loss
of androgen receptor expression predicts early recurrence in
triple-negative and basal-like breast cancer. Mod Pathol.
27:352–360. 2014.PubMed/NCBI
|
|
39
|
Tsang JY, Ni YB, Chan SK, Shao MM, Law BK,
Tan PH and Tse GM: Androgen receptor expression shows distinctive
significance in ER positive and negative breast cancers. Ann Surg
Oncol. 21:2218–2228. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Pistelli M, Caramanti M, Biscotti T,
Santinelli A, Pagliacci A, De Lisa M, Ballatore Z, Ridolfi F,
Maccaroni E, Bracci R, et al: Androgen receptor expression in early
triple-negative breast cancer: clinical significance and prognostic
associations. Cancers (Basel). 6:1351–1362. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
McNamara KM, Yoda T, Takagi K, Miki Y,
Suzuki T and Sasano H: Androgen receptor in triple negative breast
cancer. J Steroid Biochem Mol Biol. 133:66–76. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Gasparini P, Fassan M, Cascione L, Guler
G, Balci S, Irkkan C, Paisie C, Lovat F, Morrison C, Zhang J, et
al: Androgen receptor status is a prognostic marker in non-basal
triple negative breast cancers and determines novel therapeutic
options. PLoS One. 9:e885252014. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Loibl S, Müller BM, von Minckwitz G,
Schwabe M, Roller M, Darb-Esfahani S, Ataseven B, du Bois A,
Fissler-Eckhoff A, Gerber B, et al: Androgen receptor expression in
primary breast cancer and its predictive and prognostic value in
patients treated with neoadjuvant chemotherapy. Breast Cancer Res
Treat. 130:477–487. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Micello D, Marando A, Sahnane N, Riva C,
Capella C and Sessa F: Androgen receptor is frequently expressed in
HER2-positive, ER/PR-negative breast cancers. Virchows Arch.
457:467–476. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Ogawa Y, Hai E, Matsumoto K, Ikeda K,
Tokunaga S, Nagahara H, Sakurai K, Inoue T and Nishiguchi Y:
Androgen receptor expression in breast cancer: relationship with
clinicopathological factors and biomarkers. Int J Clin Oncol.
13:431–435. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Witzel I, Graeser M, Karn T, Schmidt M,
Wirtz R, Schütze D, Rausch A, Jänicke F, Milde-Langosch K and
Müller V: Androgen receptor expression is a predictive marker in
chemotherapy-treated patients with endocrine receptor-positive
primary breast cancers. J Cancer Res Clin Oncol. 139:809–816. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Tang D, Xu S, Zhang Q and Zhao W: The
expression and clinical significance of the androgen receptor and
E-cadherin in triple-negative breast cancer. Med Oncol. 29:526–533.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Hu R, Dawood S, Holmes MD, Collins LC,
Schnitt SJ, Cole K, Marotti JD, Hankinson SE, Colditz GA and Tamimi
RM: Androgen receptor expression and breast cancer survival in
postmenopausal women. Clin Cancer Res. 17:1867–1874. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Lu S, Singh K, Mangray S, Tavares R, Noble
L, Resnick MB and Yakirevich E: Claudin expression in high-grade
invasive ductal carcinoma of the breast: correlation with the
molecular subtype. Mod Pathol. 26:485–495. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Gerhard R, Ricardo S, Albergaria A, Gomes
M, Silva AR, Logullo ÂF, Cameselle-Teijeiro JF, Paredes J and
Schmitt F: Immunohistochemical features of claudin-low intrinsic
subtype in metaplastic breast carcinomas. Breast. 21:354–360. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Sabatier R, Guille A, Adelaide J,
Chaffanet M, Viens P, Birnbaum D, Bertucci F and Finetti P:
Claudin-low breast cancers: clinical, pathological, molecular and
prognostic characterization. Mol Cancer. 13:2282014. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Masuda H, Baggerly KA, Wang Y, Zhang Y,
Gonzalez-Angulo AM, Meric-Bernstam F, Valero V, Lehmann BD,
Pietenpol JA, Hortobagyi GN, et al: Differential response to
neoadjuvant chemotherapy among 7 triple-negative breast cancer
molecular subtypes. Clin Cancer Res. 19:5533–5540. 2013. View Article : Google Scholar : PubMed/NCBI
|