Introduction
Epidermal growth factor receptor (EGFR), also known
as ErbB1, is a 170-kDa transmembrane glycoprotein belonging to the
ErbB/human epidermal growth factor receptor family of receptor
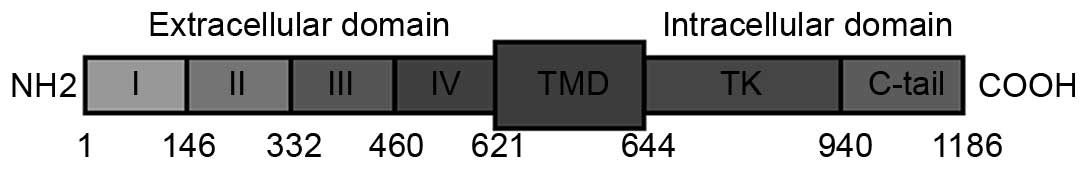
tyrosine kinases (1–3). EGFR is composed of an extracellular
highly glycosylated ligand-binding domain (ECD) comprising amino
acids 1–621, a hydrophobic transmembrane domain (amino acids
622–644) and an intracellular domain with tyrosine kinase activity
for signal transduction (amino acids 645–1,186) (Fig. 1) (1–3). Upon
binding of a ligand-like amphiregulin, EGF or transforming growth
factor α (TGFα) undergoes a conformational change by
homo-dimerization or hetero-dimerization with another member of the
erbB family, followed by auto-phosphorylation (4,5). This
results in tyrosine kinase activation and triggering of signaling
cascades. Activation of EGFR leads to the activation of
intracellular signaling pathways that regulate cell proliferation,
invasion, angiogenesis and metastasis (4,5). EGFR has
been selected as a target of anticancer treatments due to its
critical roles in cell survival and proliferation (6). EGFR is a strong prognostic marker in
head and neck, ovarian and cervical cancer (7–9). EGFR
expression has been associated with a higher proliferative index,
advanced tumor stage and increased tumor angiogenesis in HNSCC
(9). Overexpression of EGFR and TGFα
significantly predicted a shorter disease-free and overall survival
(9). EGFR activation also resulted
into increased cell invasiveness and motility (10) via the induction of
epithelial-to-mesenchymal transition (11,12).
Furthermore, EGFR can interact with the receptor cluster of
differentiation 44, resulting in a migratory cell phenotype
(13). In addition to membrane-bound
EGFR, tumor cells express soluble EGFR proteins that can be
produced by alternative messenger (m)RNA splicing events, aberrant
translocation or disintegration of circulating tumour cells
(14,15). Another 110-kDa soluble EGFR isoform,
termed proteolytic isoform-soluble (PI-s)EGFR, is disengaged by
proteolytic cleavage partially caused by metalloproteases (16,17).
Sanderson et al (18) have
also reported two soluble isoforms of EGFR (150 and 100-kDa) within
exosomes.
The present study focused on plasmatic EGFR levels
of HNSCC patients, which were analyzed by enzyme-linked
immunosorbent assay (ELISA) using anti-EGFR antibodies raised
against the L25-S645 region of full-length EGFR. Notably,
information about binding sites of ELISA antibodies are often not
provided in the literature, despite that it could be very important
for interpretation of the results obtained. Blood markers are less
invasive than tissue biopsies, and sample collection can be
repeated, which enables real-time monitoring of disease progression
and treatment response in patients. As a control group, a gender-
and age-matched healthy cohort, and a gender- and age-matched
cohort of patients with type 2 diabetes mellitus (T2DM), were used.
The T2DM group was included because a proportion of the present
HNSCC patients also exhibited T2DM, and certain studies have shown
that diabetes suppresses the expression of EGFR (19). Since EGFR is affected by both female
estrogen receptors (20,21) and male androgen receptors (22), EGFR may be a potential mediator of
gender-related differences in HNSCC. Based on these facts, female
HNSCC patients were excluded from the current study.
Materials and methods
Samples preparation
The present study was approved by the ethical
committee of St. Anne's Faculty Hospital (Brno, Czech Republic).
All surgical tissue samples were obtained from male HNSCC patients
treated at St. Anne's Faculty Hospital between April 2013 and June
2015 upon providing informed consent. Histologically verified
primary HNSCC carcinoma tissues were collected (92 samples). The
tissue material harvested at surgery was placed into
RNAlater® solution for RNA stabilization and storage
(Ambion; Thermo Fisher Scientific, Inc., Waltham, MA, USA). The
material was maintained cold, and RNA was isolated within 24 h.
Additional information about the patients and controls is presented
in Table I.
 | Table I.Characterization of patients and
controls. |
Table I.
Characterization of patients and
controls.
| Group | Factor | Number of cases | Age, years
(range) |
|---|
| HNSCC patients |
| 92 | 62.90 (44–89) |
|
| TNM T1-2 | 39 | 62.42 (44–89) |
|
| TNM T3-4 | 52 | 63.17 (47–87) |
|
| TNM N0 | 42 | 65.04 (44–89) |
|
| TNM N1 | 49 | 61.26 (44–77) |
|
| TNM M0 | 86 | 62.86 (44–89) |
|
| TNM M1 | 5 | 62.12 (55–71) |
|
| Grade 1 | 6 | 63.46 (53–79) |
|
| Grade 2 | 50 | 63.26 (44–89) |
|
| Grade 3 | 32 | 61.49 (47–75) |
|
| Non-smoker | 28 | 66.99 (46–89) |
|
| Smoker | 59 | 62.01 (44–78) |
| Healthy
controls |
| 29 | 64.38 (54–69) |
| Diabetic
controls |
| 26 | 56.73 (50–83) |
Blood samples from HNSCC patients and healthy (N=29)
and T2DM (N=26) controls were obtained by venipuncture, and 5 ml
was placed into an S-Monovette® 4.9 ml, K3EDTA test-tube
(Sarstedt AG & Co., Nümbrecht, Germany) for plasma preparation.
The blood samples were centrifuged at 1,200 × g at 4°C for 10 min
within 60 min of collection. Plasma was aliquoted and stored at
−80°C until analysis.
ELISA analysis
Plasma levels of EGFR were determined with a
commercial ELISA kit (RayBiotech, Inc., Norcross, GA, USA)
according to the manufacturer's protocol. The ELISA was designed to
detect human EGFR in plasma or serum with a detection limit of 4
pg/ml, a 10% intra-assay varaibility and a 12% inter-assay
variability, as described in the manufacturer's instructions. For
the assay, plasma samples were diluted 100-fold, and evaluated with
anti-EGFR antibodies raised against the L25-S645 region of
EGFR.
RNA isolation and reverse
transcription (RT)
TriPure Isolation reagent (Roche Diagnostics, Basel,
Switzerland) was used for RNA isolation. The isolated RNA was used
for complementary (c)DNA synthesis. RNA (1,000 ng) was reverse
transcribed using Transcriptor First Strand cDNA Synthesis kit
(Roche Diagnostics) according to manufacturer's protocol. The cDNA
(20 µl) prepared from total RNA was diluted with RNase-free water
to 100 µl, and 5 µl cDNA was directly analyzed using the
LightCycler® 480 II System (Roche Diagnostics).
RT-quantitative polymerase chain
reaction (qPCR)
RT-qPCR was performed using TaqMan® Gene
Expression Assays (Life Technologies; Thermo Fisher Scientific,
Inc.) with the LightCycler® 480 II System, and the
amplified DNA was analyzed by the comparative ΔΔCq calculation
(23) using β-actin as an endogenous
control. The primer and probe sets for β-actin (Hs99999903_m1),
metallothionein (MT)2 (Hs02379661_g1), MT1 (Hs00831826_s1), tumor
protein p53 (TP53) (Hs01034249_m1), B-cell lymphoma (BCL)-2
associated X protein (BAX) (Hs00180269_m1), BCL-2 (Hs00608023_m1),
vascular endothelial growth factor A (VEGFA) (Hs00900055_m1),
fms-related tyrosine kinase 1 (FLT1) (Hs01052961_m1), matrix
metalloproteinase 2 (MMP2) (Hs01548727_m1), MMP9 (Hs00234579_m1),
proto-oncogene c-Fos (FOS) (Hs00170630_m1), c-Jun (JUN)
(Hs00277190_s1), marker of proliferation Ki-67 (MKI67)
(Hs00606991_m1), EGF (Hs01099999_m1) and EGFR (Hs01076078_m1) were
selected from TaqMan® Gene Expression Assays. RT-qPCR
was performed under the following amplification conditions in a
total volume of 20 µl (5 µl cDNA, 10 µl TaqMan® Gene
Expression Master Mix, 4 µl molecular-grade water and 1 µl TaqMan
Gene Expression Assay): Initial incubation, 50°C for 2 min,
followed by denaturation at 95°C for 10 min and then 45 cycles of
95°C for 15 sec and 60°C for 1 min.
Human papillomavirus (HPV)
detection
The 142 bp-long sequence of the conservative major
capsid protein L1 gene were amplified using general primers, GP5
and GP6, for non-specific identification of HPV-positive subjects.
The PCR mixture from New England BioLabs, Inc. (Ipswich, MA, USA)
contained PCR buffer (10 mM Tris HCl, pH 8.3, 50 mM KCl and 2.5 mM
MgCl2), 0.05 mM of each deoxynucleotide, and 0.05 mM of
GP5 (5′-TTTGTTACTGTGGTAGATAC-3′) and GP6
(5′-GAAAAATAAACTGTAAATCA-3′) primers. The DNA amplification was
performed during 40 cycles that included a denaturation step at
94°C for 30 sec, annealing at 45°C for 30 sec and extension at 72°C
for 30 sec.
As internal quality control of the isolated DNA, the
β-actin gene (600 bp) was amplified (forward primer
5′-CCTGAACCCTAAGGCCAACC-3′ and reverse primer
5′-GCAATGCCTGGGTACATGGT-3′). Each PCR product was analyzed using
electrophoresis on 1% agarose gels stained with ethidium
bromide.
Data analysis
Differences between the two groups were calculated
using the t-test. Survival analysis was conducted using Cox
proportional hazard regression analysis with plasma EGFR levels as
covariates. Receiver operating characteristic (ROC) curves were
calculated using the DeLong methodology. Subsequently, Kaplan-Meier
analysis was used with continuous data being divided into two
groups as follows: Low expression (<mean values) and high
expression (≥mean values) groups. The associations between the
continuous variables were analyzed using Pearson's correlations.
Unless noted otherwise, P<0.05 was considered to indicate a
statistically significant difference. Software STATISTICA 12
(StatSoft, Inc., Tulsa, OK, USA) and MedCalc 15.8 (MedCalc Software
bvba, Ostend, Belgium) were used for analysis.
Results
Association between plasma levels of
EGFR and HNSCC occurrence
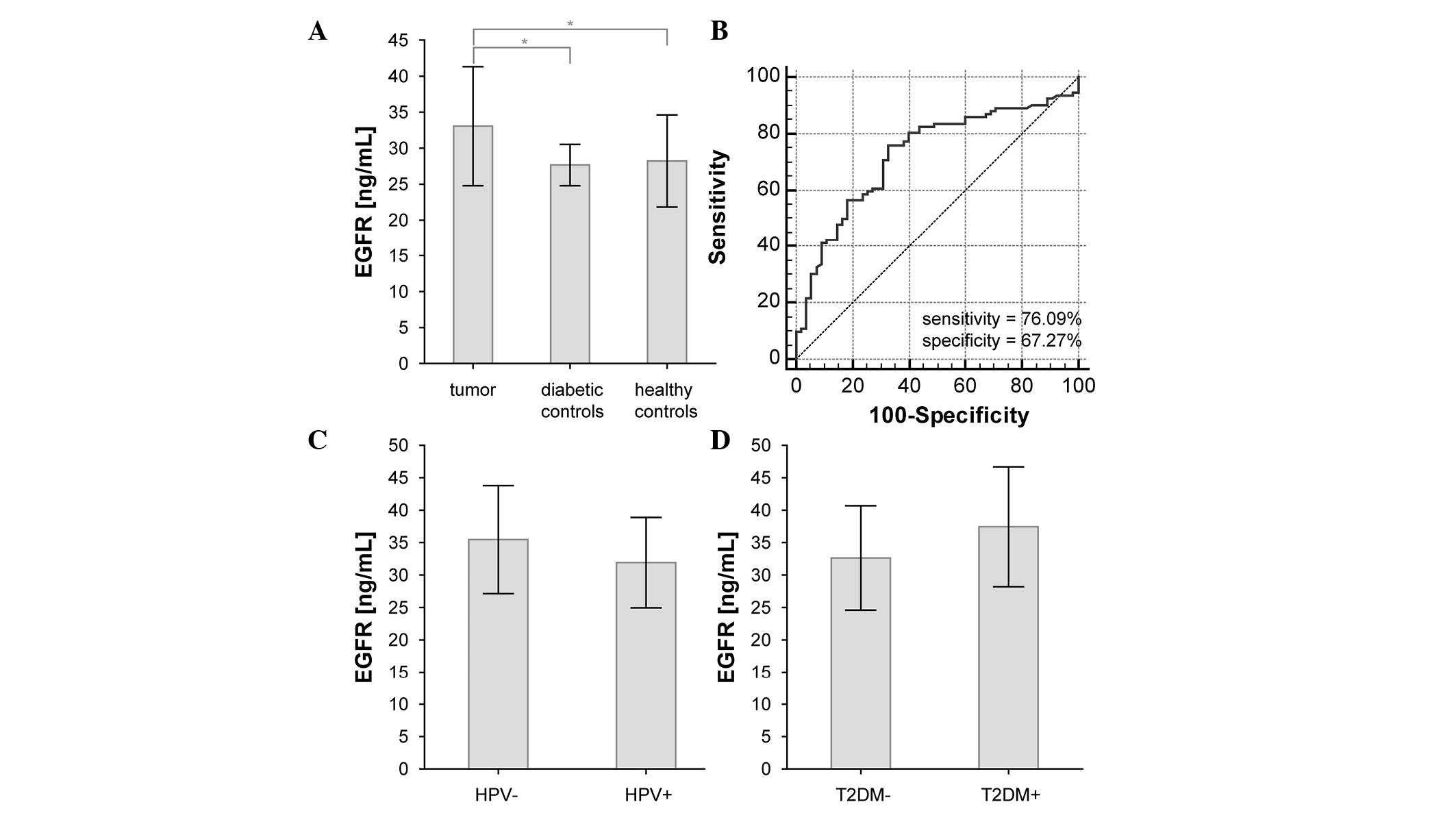
No significant changes in EGFR plasma levels were
observed between diabetic and healthy controls (P=0.690). However,
there was a significant difference between EGFR plasma levels in
HNSCC patients and in both control groups (P=0.001 and 0.005,
respectively) (Fig. 2A and Table II). If both control groups were
assessed together, the statistical significance was P=0.0001. ROC
curve analysis identified a sensitivity of 76.09%, a specificity of
67.27% and an area under the curve (AUC) of 0.727 for this
comparison (Fig. 2B). Additional
information about the patients and controls is contained in
Table I.
 | Table II.Plasma levels of EGFR and clinical
characteristics in HNSCC patients. |
Table II.
Plasma levels of EGFR and clinical
characteristics in HNSCC patients.
| Factor | Status (number of
cases) | EGFR levels, ng/ml
mean ± standard deviation | P-value |
|---|
| Cases vs.
controls | HNSCC patients
(92) | 33.1±8.3 | – |
|
| Healthy controls
(29) | 28.2±6.4 | 0.001 |
|
| Diabetic controls
(26) | 27.7±2.9 | 0.005 |
| Smoking | Yes (59) | 31.8±7.7 | 0.150 |
|
| No (28) | 34.5±8.5 | – |
| Hypertension | Yes (28) | 34.4±8.8 | 0.380 |
|
| No (58) | 32.7±8.0 | – |
| Diabetes
mellitus | Yes (10) | 37.4±9.2 | 0.085 |
|
| No (76) | 32.6±8.0 | – |
| TNM T-staging | T1-2 (39) | 32.5±8.1 | 0.580 |
|
| T3-4 (52) | 33.4±8.5 | – |
| TNM N-staging | N+ (49) | 33.7±8.2 | 0.430 |
|
| N- (42) | 32.3±8.5 | – |
| TNM M-staging | M+ (5) | 31.3±8.1 | 0.640 |
|
| M- (86) | 33.1±8.3 | – |
| Tumor grade | High (82) | 32.8±8.1 | 0.270 |
|
| Low (6) |
36.8±12.3 | – |
| HPV status | HPV+ (49) | 32.0±7.0 | 0.084 |
|
| HPV- (18) | 35.5±8.3 | – |
Correlation between tumor gene
expression and EGFR plasma levels
Correlations between plasma EGFR levels and
expression of genes in tumor tissues of HNSCC patients were
examined. There was no significant correlation between plasma EGFR
levels and tumor tissue EGFR mRNA expression, and only a weak
negative correlation with MMP9 mRNA was observed (r=−0.21,
P=0.040). Gene expression analyses of EGF, EGFR, MKI67, BCL-2, BAX,
FOS, JUN, TP53, VEGFA, FLT1, MMP2, MMP9, MT1A and MT2A genes in
HNSCC tumor tissue compared with tumor adjacent tissue and
tonsillectomies have been published elsewhere (24).
Association between plasma levels of
EGFR and clinicopathological characteristics
By examining the associations between plasma EGFR
levels and clinicopathological characteristics of HNSCC patients,
no significant association was identified for smoking habit, T2DM,
hypertension, HPV infection, tumor stage, tumor grade, or lymph
node or distant metastasis occurrence. However, the presence of HPV
infection and T2DM in HNSCC patients had a borderline effect on the
plasma EGFR levels (Table II and
Fig. 2C and D).
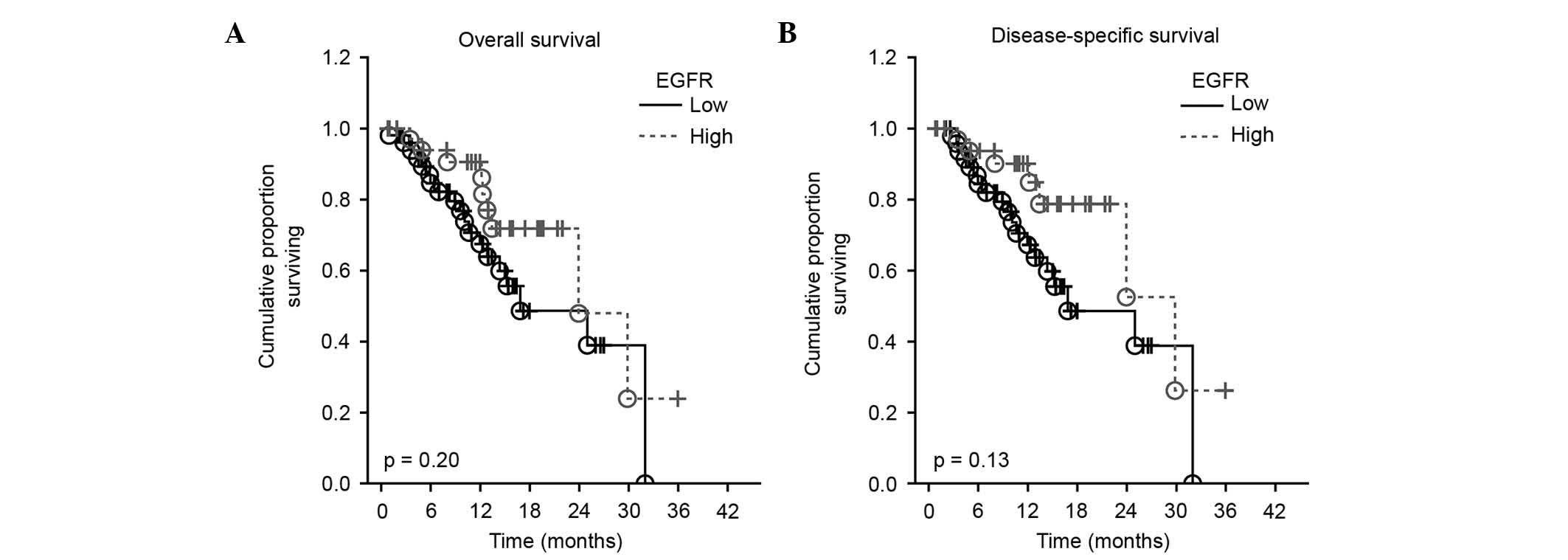
Association between plasma levels of
EGFR and disease-free and overall survival
The prognostic value of EGFR plasma levels on
overall and disease-free survival was studied by Cox proportional
hazard regression analysis and Kaplan-Meier curves. Survival
analysis revealed no significant influence of plasmatic EGFR levels
on overall or disease-specific survival in the present cohort of
HNSCC patients [hazard ratio (HR)=0.97; 95% confidence interval
(CI)=0.92–1.01; P=0.200, and HR=0.96; 95 CI=0.91–1.01; P=0.130 for
overall and disease-free survival, respectively] (Fig. 3).
Discussion
Numerous studies have shown that EGFR is
overexpressed in HNSCC tumor tissue, but only few studies focused
on soluble EGFR levels (24–26). There are contradictory studies on
soluble EGFR levels, which could be either decreased or elevated in
cancer patients compared with a healthy cohort. For example,
Partanen et al reported that patients with
asbestosis-induced lung cancer have elevated serum soluble EGFR ECD
levels (27). Increased soluble EGFR
ECD levels were also reported in the urine of patients with
squamous cell carcinomas of the lung, head and neck (28), whereas patients with ovarian cancer
had decreased levels of serum p110 EGFR compared with the normal
population (29). In the present
study, ELISA using antibodies against the L25-S645 region of EGFR
was used to measure the levels of EGFR in plasma samples of HNSCC
patients. Female patients were excluded from the present study due
to possible gender-specific EGFR interactions with estrogen or
androgen receptors (20–22). Significantly higher EGFR plasma levels
were detected in HNSCC patients compared with the healthy cohort
and the diabetic control group (P=0.001 and 0.005, respectively).
This finding is in accordance with that of Perez-Torres et
al (17), who suggested that the
mechanism of proteolytic cleavage of EGFR and shedding of PI-sEGFR
into the plasma may be activated in malignant cells that
overexpress the full-length receptor. The cleavage of EGFR probably
occurs in the transmembrane domain between G625 and M626 (17). In HNSCC patients, EGFR expression is
supposed to be higher in tumor tissues compared with tonsillectomy
samples and tumor-adjacent tissues (24). Furthermore, the release of two soluble
EGFR isoforms within the exosomes is activated by EGF (17), which is highly produced by HNSCC
tumor-adjacent tissues (24).
No significant changes in EGFR plasma levels were
observed between diabetic and healthy controls, which is not in
accordance with the Vairaktaris et al hypothesis that
diabetes suppresses the expression of EGFR (19). However, a slight change on the
borderline of statistical significance was observed between HNSCC
patients with or without diabetes (P=0.085), while HNSCC patients
with diabetes tended to have higher EGFR plasma levels. Borderline
changes in EGFR plasma levels were also noticed between the
HPV-positive and HPV-negative groups of HNSCC patients (slightly
higher levels of plasmatic EGFR were detected in the HPV-negative
cohort), although these changes were not significant.
Survival analysis revealed no significant influence
of the plasmatic EGFR levels on overall and disease-specific
survival in the present cohort of HNSCC patients. By contrast, Ye
et al demonstrated that non-small-cell lung cancer patients
with lower plasma EGFR concentrations (<27.24 ng/ml) had a
significantly shorter overall survival compared with patients who
had higher plasma EGFR concentrations (≥27.24 ng/ml) (18.2 vs. 33.4
months, P=0.021) (30).
In conclusion, EGFR plasma levels appear to be a
relatively promising diagnostic, but poor prognostic, HNSCC marker.
However, further studies are required to determine the clinical
value of plasmatic EGFR levels in HNSCC patients. The next
important step in soluble EGFR research should be a precise
distinction between 110-kDa PI-sEGFR originating from full-length
EGFR protein cleavage and EGFR isoforms originating from
alternative splicing of EGFR gene transcripts. These EGFR isoforms
could readily have slightly different functions. For example,
110-kDa PI-sEGFR originating from full-length EGFR protein cleavage
could reflect the presence of malignant cells that overexpress the
full-length receptor (17) or a
necrotic disintegration of tumor cells. Such form of soluble EGFR
was probably originally involved in a proliferative signaling
pathway, and could be marker of poor prognosis, while the soluble
EGFR isoform originating from alternative splicing was probably not
an activator of these proliferative signaling pathways due to the
missing intracellular domain, and could exhibit a high affinity
binding for EGF, which should result in decreased proliferative
signaling and better prognosis.
Acknowledgements
The present study was supported by the Ministry of
Health of the Czech Republic (Prague, Czech Republic; grant no. IGA
MZ NT 14337-3/2013) and by Specific University Research Grants
(grant nos. MUNI/A/1365/2015 and MUNI/A/1426/2015) provided by the
Ministry of Education, Youth and Sports of the Czech Republic
(Prague, Czech Republic) in 2016 and by Czech Science Foundation
(GACR GA16-12454S).
References
|
1
|
Albitar L, Pickett G, Morgan M, Wilken JA,
Maihle NJ and Leslie KK: EGFR isoforms and gene regulation in human
endometrial cancer cells. Mol Cancer. 9:1662010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ullrich A, Coussens L, Hayflick JS, Dull
TJ, Gray A, Tam AW, Lee J, Yarden Y, Libermann TA, Schlessinger J,
et al: Human epidermal growth-factor receptor cDNA sequence and
aberrant expression of the amplified gene in A431 epidermoid
carcinoma-cells. Nature. 309:418–425. 1984. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ciardiello F and Tortora G: Epidermal
growth factor receptor (EGFR) as a target in cancer therapy:
Understanding the role of receptor expression and other molecular
determinants that could influence the response to anti-EGFR drugs.
Eur J Cancer. 39:1348–1354. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Psyrri A, Seiwert TY and Jimeno A:
Molecular pathways in head and neck cancer: EGFR, PI3K, and more.
Am Soc Clin Oncol Educ Book. 246–255. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Sartor CI: Biological modifiers as
potential radiosensitizers: Targeting the epidermal growth factor
receptor family. Semin Oncol. 27(6 Suppl 11): S15–S20; discussion
92–100. 2000.
|
|
6
|
Woodburn JR: The epidermal growth factor
receptor and its inhibition in cancer therapy. Pharmacol Ther.
82:241–250. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Nicholson RI, Gee JM and Harper ME: EGFR
and cancer prognosis. Eur J Cancer. 37(Suppl 4): S9–S15. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Salomon DS, Brandt R, Ciardiello F and
Normanno N: Epidermal growth factor-related peptides and their
receptors in human malignancies. Crit Rev Oncol Hematol.
19:183–232. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Grandis Rubin J, Melhem MF, Gooding WE,
Day R, Holst VA, Wagener MM, Drenning SD and Tweardy DJ: Levels of
TGF-alpha and EGFR protein in head and neck squamous cell carcinoma
and patient survival. J Natl Cancer Inst. 90:824–832. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Box C, Rogers SJ, Mendiola M and Eccles
SA: Tumour-microenvironmental interactions: Paths to progression
and targets for treatment. Semin Cancer Biol. 20:128–138. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zuo JH, Zhu W, Li MY, Li XH, Yi H, Zeng
GQ, Wan XX, He QY, Li JH, Qu JQ, et al: Activation of EGFR promotes
squamous carcinoma SCC10A cell migration and invasion via inducing
EMT-Like phenotype change and MMP-9-mediated degradation of
E-cadherin. J Cell Biochem. 112:2508–2517. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Holz C, Niehr F, Boyko M, Hristozova T,
Distel L, Budach V and Tinhofer I:
Epithelial-mesenchymal-transition induced by EGFR activation
interferes with cell migration and response to irradiation and
cetuximab in head and neck cancer cells. Radiother Oncol.
101:158–164. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wang SJ and Bourguignon LY: Hyaluronan and
the interaction between CD44 and epidermal growth factor receptor
in oncogenic signaling and chemotherapy resistance in head and neck
cancer. Arch Otolaryngol Head Neck Surg. 132:771–778. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hunts JH, Shimizu N, Yamamoto T, Toyoshima
K, Merlino GT, Xu YH and Pastan I: Translocation chromosome 7 of
A431 cells contains amplification and rearrangement of EGF receptor
gene responsible for production of variant mRNA. Somat Cell Mol
Genet. 11:477–484. 1985. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kulasinghe A, Perry C, Jovanovic L, Nelson
C and Punyadeera C: Circulating tumour cells in metastatic head and
neck cancers. Int J Cancer. 136:2515–2523. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ancot F, Foveau B, Lefebvre J, Leroy C and
Tulasne D: Proteolytic cleavages give receptor tyrosine kinases the
gift of ubiquity. Oncogene. 28:2185–2195. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Perez-Torres M, Valle BL, Maihle NJ,
Negron-Vega L, Nieves-Alicea R and Cora EM: Shedding of epidermal
growth factor receptor is a regulated process that occurs with
overexpression in malignant cells. Exp Cell Res. 314:2907–2918.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Sanderson MP, Keller S, Alonso A, Riedle
S, Dempsey PJ and Altevogt P: Generation of novel, secreted
epidermal growth factor receptor (EGFR/ErbB1) isoforms via metal
loprotease-dependent ectodomain shedding and exosome secretion. J
Cell Biochem. 103:1783–1797. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Vairaktaris E, Goutzanis L, Yapijakis C,
Vassiliou S, Spyridonidou S, Vylliotis A, Nkenke E, Lazaris AC,
Strantzias P and Patsouris E: Diabetes enhances the expression of
H-ras and suppresses the expression of EGFR leading to increased
cell proliferation. Histol Histopathol. 24:531–539. 2009.PubMed/NCBI
|
|
20
|
Britton DJ, Hutcheson IR, Knowlden JM,
Barrow D, Giles M, McClelland RA, Gee JM and Nicholson RI:
Bidirectional cross talk between ERalpha and EGFR signalling
pathways regulates tamoxifen-resistant growth. Breast Cancer Res
Treat. 96:131–146. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Levin ER: Bidirectional signaling between
the estrogen receptor and the epidermal growth factor receptor. Mol
Endocrinol. 17:309–317. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Bonaccorsi L, Muratori A, Carloni V,
Marchiani S, Formigli L, Forti G and Baldi E: The androgen receptor
associates with the epidermal growth factor receptor in
androgen-sensitive prostate cancer cells. Steroids. 69:549–552.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Raudenska M, Sztalmachova M, Gumulec J,
Fojtu M, Polanska H, Balvan J, Feith M, Binkova H, Horakova Z,
Kostrica R, et al: Prognostic significance of the tumour-adjacent
tissue in head and neck cancers. Tumour Biol. 36:9929–9939. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Polanska H, Raudenska M, Gumulec J,
Sztalmachova M, Adam V, Kizek R and Masarik M: Clinical
significance of head and neck squamous cell cancer biomarkers. Oral
Oncol. 50:168–177. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Grandis JR and Tweardy DJ: Elevated levels
of transforming growth-factor-alpha and epidermal growth-factor
receptor messenger RNA are early markers of carcinogenesis in head
and neck-cancer. Cancer Res. 53:3579–3584. 1993.PubMed/NCBI
|
|
27
|
Partanen R, Hemminki K, Koskinen H, Luo
JC, Carney WP and Brandtrauf PW: The detection of increased amounts
of the extracellular domain of the epidermal growth-factor receptor
in serum during carcinogenesis in asbestosis patients. J Occup Med.
36:1324–1328. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Witters LM, Curley EM, Kumar R, Chinchilli
VM, Harvey JP, Crebbin V, Harvey HA and Lipton A: Epidermal growth
factor receptor ectodomain in the urine of patients with squamous
cell carcinoma. Clin Cancer Res. 1:551–557. 1995.PubMed/NCBI
|
|
29
|
Carney WP: Circulating oncoproteins
HER2/neu, EGFR and CAIX (MN) as novel cancer biomarkers. Expert Rev
Mol Diagn. 7:309–319. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Ye P, Zhao J, Wang S and Kong FM: The
plasma level of soluble epidermal growth factor Receptor (EGFR) and
overall survival (OS) in non-small-cell lung cancer (NSCLC)
patients. Annual Meeting of the American Society of Clinical
Oncology (ASCO). J Clin Oncol. 33:e190912015.
|