Networks analysis of genes and microRNAs in human Wilms' tumors
- Authors:
- Published online on: September 7, 2016 https://doi.org/10.3892/ol.2016.5102
- Pages: 3579-3585
Metrics: Total
Views: 0 (Spandidos Publications: | PMC Statistics: )
Total PDF Downloads: 0 (Spandidos Publications: | PMC Statistics: )
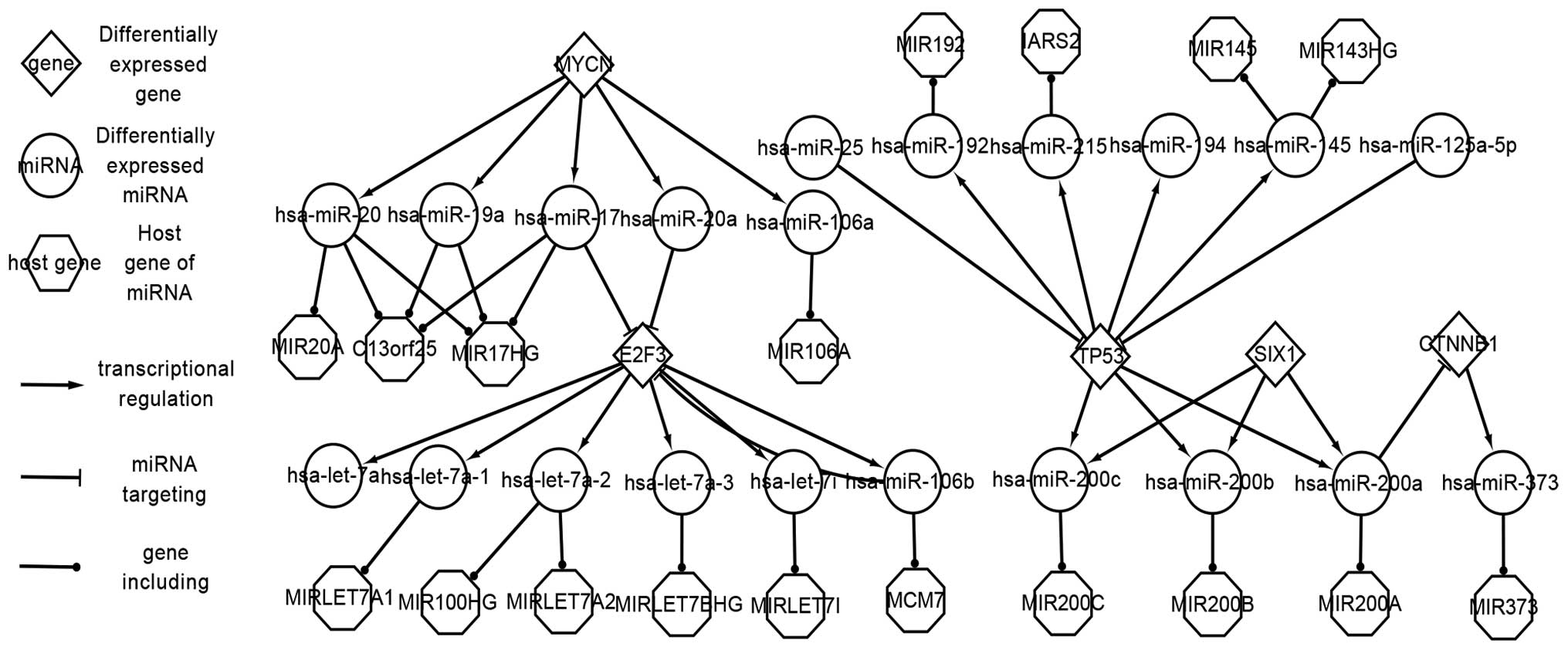
Abstract
Wilms' tumor (WT) is a common kidney cancer. To date, the expression of genes [transcription factors (TFs), target genes and host genes] and microRNAs (miRNAs/miRs) in WTs has captured the attention of biologists, while the regulatory association between the genes and miRNAs remains unclear. In the present study, TFs, miRNAs, target genes and host genes were considered as key factors in the construction of three levels of regulatory networks, namely, the differentially‑expressed network, the related network and the global network. The four factors had three types of association, including the regulation of miRNAs by TFs, the targeting of the target genes by miRNAs and the location of miRNAs at host genes. The differentially-expressed network is the most important of the three networks, and only involves the differentially‑expressed genes and miRNAs; with the exception of host genes, those elements all behave abnormally when a WT occurs, which suggests that the differentially‑expressed network can accurately reveal the pathogenesis of WTs. E2F3, for example, is overexpressed in WTs, and it regulates hsa‑let‑7a, hsa‑let‑7a‑1, hsa‑miR‑106b, among others. Meanwhile, E2F3 is targeted by hsa‑miR‑106b, hsa‑miR‑17 and hsa‑miR‑20a. If the regulatory network can be used to adjust those factors to a normal level, there may be a chance to cure patients with WTs. WT‑associated factors were placed into the related network; this network is useful for understanding the regulatory pathways of genes and miRNA in WTs. The networks provide a novel perspective in order to study the inner interactions of genes and miRNAs. The present study provides authoritative data and regulatory pathway analysis in order to partially elucidate the pathogenesis of WT, and thus supplies biologists with a basis for future research.