Introduction
Breast cancer, which accounts for ~23% of all newly
diagnosed cases of cancer and was responsible for 14% (458,400) of
all mortalities due to cancer in 2008, is the leading cause of
cancer-associated mortality among females (1). A previous study reported that the
estrogen receptor (ER), which is expressed in ~75% of breast
tumors, is considered the main target for the treatment of breast
cancer, and women with breast tumors typically receive endocrine
therapy (2).
Fulvestrant, which is a pure, steroidal
antiestrogen, has been reported to completely suppress ERα activity
by inactivating ERα-mediated genomic and non-genomic signaling; it
is considered a promising drug for the treatment of breast cancer
in postmenopausal women (3). However,
ER-targeted therapies fail in ≤50% of patients with breast tumors
due to the occurrence of de novo or acquired resistance
(2,4).
It has been reported that microRNAs (miRNAs) have a pivotal role in
breast cancer, and the overexpression of miRNA-221/222 has been
suggested to be associated with the emergence of fulvestrant
resistance in breast cancer (5).
In 2011, Rao et al (6) used a microarray expression profile to
identify differentially expressed genes (DEGs) between antisense
miRNA-221-transfected or miRNA-222-transfected MCF7-FR cells and
negative control-transfected MCF7-FR cells, according to the
cut-off criteria of P<0.05 and |log2 fold change
(FC)| >1.2. It was demonstrated that activation of β-catenin by
miRNA-221/222 led to estrogen-independent growth and fulvestrant
resistance, as well as to repression of transforming growth
factor-β-mediated growth inhibition (6). However, another study reported different
mechanisms for the occurrence of fulvestrant resistance in breast
cancer (7). Tangkeangsirisin and
Serrero (8) demonstrated that
progranulin induced human breast cancer resistance to fulvestrant
by inhibiting the apoptosis of breast cancer cells. In addition,
the broad-spectrum metalloproteinase inhibitor BB-94 has been
demonstrated to inhibit the growth of fulvestrant-resistant breast
cancer cell lines, as well as the activation of human epidermal
growth factor receptor 3 and extracellular signal-regulated kinase
in these cells (9). Therefore, it is
important to further screen for biomarkers associated with
fulvestrant-resistance in breast cancer.
Using the same microarray data as Rao et al
(6), the present study aimed to
further screen for DEGs in antisense miRNA-221-transfected and
antisense miRNA-222-transfected MCF7-FR cells. The linear models
for microarray data (limma) package, based on a wide threshold
range (P<0.05 and |log2 FC| >1), was used to
identify DEGs associated with fulvestrant-resistant breast cancer.
In addition, a Kyoto Encyclopedia of Genes and Genomes (KEGG)
pathway enrichment analysis was performed, and the targets of
miRNA-221/222 were predicted using miRanda and TargetScan. A
previous study suggested that analyses based on different
statistical tests may produce different outcomes (10). Therefore, the present study may obtain
a number of results different from the data obtained in the initial
study by Rao et al (6).
Materials and methods
Microarray data
The GSE19777 transcription profile used by Rao et
al (6) was downloaded from the
Gene Expression Omnibus database (http://www.ncbi.nlm.nih.gov/geo/). The profile was
based on the GPL570 dataset, which was obtained using the
[HG-U133_Plus_2] Affymetrix Human Genome U133 Plus 2.0 Array
(Affymetrix, Inc., Santa Clara, CA, USA). In total, nine samples
were included in the dataset, including three samples of antisense
miRNA-221-transfected fulvestrant-resistant MCF7-FR breast cancer
cells, three samples of antisense miRNA-222-transfected
fulvestrant-resistant MCF7-FR cells and three samples of control
inhibitor (green fluorescent protein)-treated fulvestrant-resistant
MCF7-FR cells (negative control). In addition, the probe annotation
information mapping the probes of genes was downloaded from
Bioconductor (http://www.bioconductor.org/).
Dataset preprocessing and DEG
analysis
The R package from Affymetrix, Inc., was used to
normalize the raw CEL data from the DNA microarrays (11). The downloaded expression profile was
mapped to the corresponding gene symbols. Average expression values
were used for the genes with multiple probes. Subsequently, the
limma package in R/Bioconductor (https://bioconductor.org/packages/release/bioc/html/limma.html)
was used to screen for DEGs in the antisense miRNA-221-transfected
and miRNA-222-transfected MCF7-FR cells, as compared with the
negative control. The cut-off criteria for the DEGs were P<0.05
and |log2 FC| >1. The top ten upregulated and
downregulated genes in the antisense miRNA-221-transfected and
antisense miRNA-222-transfected MCF7-FR cells are indicated in
Table I. Next, the pheatmap package
(https://cran.r-project.org/web/packages/pheatmap/index.html)
in R was used to perform two-way clustering (12), based on the Euclidean distance
(13).
 | Table I.Top ten upregulated and downregulated
DEGs in the antisense miRNA-221-transfected and antisense
miRNA-222-transfected MCF7-FR cells, as compared with negative
control-transfected MCF7-FR cells. |
Table I.
Top ten upregulated and downregulated
DEGs in the antisense miRNA-221-transfected and antisense
miRNA-222-transfected MCF7-FR cells, as compared with negative
control-transfected MCF7-FR cells.
|
| Antisense miRNA-221
vs. control | Antisense miRNA-222
vs. control |
|---|
|
|
|
|
|---|
| DEGs | Gene | |log2
FC| | adj.P.Val | Gene | |log2
FC| | adj.P.Val |
|---|
| Downregulated | LHX8 | −4.17614 | 0.000175 | PTH | −3.49280 | 0.000184 |
|
| PSMB8 | −4.06074 | 0.000204 | PWAR5 | −3.27792 | 0.009442 |
|
| FLG2 | −3.99026 | 0.001523 | IYD | −3.20234 | 0.001261 |
|
| TRPC5 | −3.83132 | 0.002291 | ICAM5 | −2.87824 | 0.017322 |
|
| OLFM4 | −3.51933 | 0.000522 | DAOA-AS1 | −2.83576 | 0.044589 |
|
| KERA | −3.37161 | 0.006165 |
STARD13-AS | −2.79578 | 0.005935 |
|
| GIMAP2 | −3.19819 | 0.008755 | WDR86-AS1 | −2.75129 | 0.021120 |
|
| CYP4F30P | −2.92305 | 0.042475 | IZUMO2 | −2.68134 | 0.028787 |
|
|
LOC100505635 | −2.89457 | 0.041871 | RAG2 | −2.67795 | 0.006909 |
|
| SLC15A3 | −2.88269 | 0.000007 |
C1orf192 | −2.66616 | 0.022020 |
| Upregulated | PRPS1L1 | 3.81365 | 0.002733 | OR2L13 | 3.55457 | 0.007431 |
|
|
ARHGAP36 | 3.33814 | 0.000913 | PRPS1L1 | 3.54601 | 0.002105 |
|
| CXorf58 | 3.28644 | 0.000074 |
SH3RF3-AS1 | 3.34868 | 0.000066 |
|
|
LINC00567 | 3.24582 | 0.000653 | CXorf58 | 3.15695 | 0.028128 |
|
| DYDC1 | 3.18358 | 0.026382 |
LOC100505676 | 3.11364 | 0.001263 |
|
| OR2L13 | 2.88576 | 0.003734 |
LINC00950 | 2.92266 | 0.001549 |
|
| MLIP | 2.83880 | 0.036347 | NXPH1 | 2.90566 | 0.000380 |
|
| KLKB1 | 2.79668 | 0.015902 | MSTN | 2.89563 | 0.000007 |
|
|
LINC00950 | 2.75805 | 0.001693 | DZIP1 | 2.86233 | 0.007720 |
|
| OR10A5 | 2.74849 | 0.011940 |
CPEB2-AS1 | 2.85775 | 0.015210 |
Gene set enrichment analysis
(GSEA)
GSEA, which is a computational method that
determines whether an a priori defined set of genes exhibits
statistically significant and concordant differences between two
biological states (14), was used to
conduct the pathway enrichment analysis based on the expression
levels of DEGs in the antisense miRNA-221-transfected and
miRNA-222-transfected MCF7-FR cells. A gene count between 15 and
500 and P<0.01 were set as the criteria to filter the
pre-defined gene sets. In addition, the distant regulatory elements
of co-regulated genes tool (http://dire.dcode.org), which enables the prediction
of distant regulatory elements in higher eukaryotic genomes
(15), was applied to screen for
transcription factors associated with the DEGs in the enriched
pathways.
miRNA-messenger (m) RNA regulatory
network construction
Prediction of the targets of miRNA-221 and miRNA-222
was performed using the miRanda algorithm (http://microrna.sanger.ac.uk/targets/v5/) and
TargetScan 4.2 (http://www.targetscan.org/). Subsequently, the
miRNA-mRNA regulatory network, depicting interactions between the
miRNAs and target DEGs (upregulated DEGs only) was constructed and
visualized using Cytoscape (16).
Results
Preprocessing and DEG analysis
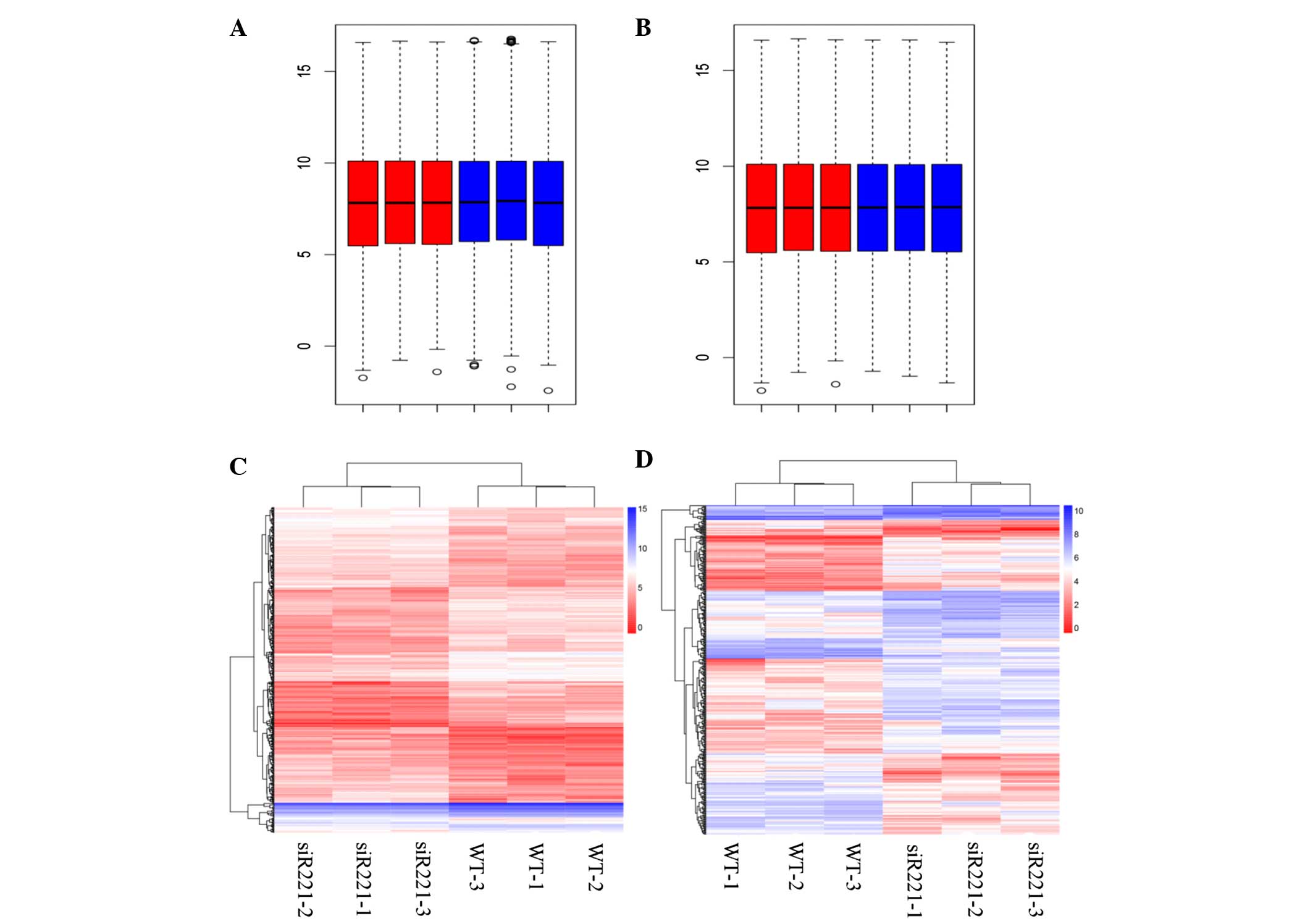
The box plots of the expression values for all genes
in every sample following normalization are represented in Fig. 1A and B. In total, 492 DEGs, including
247 upregulated [such as phosphoribosyl pyrophosphate synthetase
1-like 1 (PRPS1L1) and secreted frizzled-related protein 5
(SFRP5)] and 245 downregulated (such as LIM homeobox 8 and
proteasome subunit beta 8) DEGs, were identified in the antisense
miRNA-221-transfected MCF7-FR cells compared with the negative
control, while 404 DEGs, including 255 upregulated [such as
PRPS1L1 and claudin 8 (CLDN8)] and 149 downregulated
(such as parathyroid hormone and Prader Willi/Angelman region RNA
5) DEGs, were identified in the antisense miRNA-222-transfected
MCF7-FR cells compared with the negative control. The two-way
hierarchical cluster analyses of the DEGs in the miRNA-221- and
miRNA-222-transfected cells are represented in Fig. 1C and D.
GSEA
Three pathways were significantly enriched in the
antisense miRNA-221-transfected MCF7-FR cells compared with the
negative control, while ten pathways were significantly enriched in
the miRNA-221-transfected MCF7-FR cells compared with the negative
control (Table II). In addition, two
pathways, including the pentose phosphate pathway (PPP) and
olfactory transduction, were enriched in both the antisense
miRNA-221-transfected and miRNA-222-transfected MCF7-FR cells, as
compared with the negative control (Table II). Notably, the DEGs SFRP5
and CLDN8 were significantly enriched in the Wnt signaling
pathway and the cell adhesion molecules (CAMs) pathway,
respectively (Table II).
 | Table II.Enriched pathways for DEGs in the
miRNA-221-transfected and miRNA-222-transfected MCF7-FR cells, as
compared with the negative control-transfected MCF7-FR cells. |
Table II.
Enriched pathways for DEGs in the
miRNA-221-transfected and miRNA-222-transfected MCF7-FR cells, as
compared with the negative control-transfected MCF7-FR cells.
| Groups | Pathway | Counts | P-value | Top 10 DEGs |
|---|
| Antisense
miRNA-221-transfected MCF7-FR cells | PPP | 26 | 0 | RPE, RPIA, PGM2,
PGLS, PRPS2, FBP2, PFKM, PFKL, TALDO1, TKT |
|
| Histidine
metabolism | 27 | 0 | CNDP1, MAOB,
MAOA, ALDH1B1, ALDH2, METTL6, WBSCR22, HAL, HNMT |
|
| Olfactory
transduction | 114 | 0 | CALM2, CALM1,
OR11H4, OR52W1, OR5AU1, ADRBK2, OR2M2, OR2M7, OR2T33,
OR4F5 |
| Antisense
miRNA-222-transfected MCF7-FR cells | PPP | 26 | 0 | RPE, RPIA, PGM2,
PGLS, PRPS2, FBP2, PFKM, PFKL, TALDO1, TKT |
|
| Taste
transduction | 44 | 0 | TAS2R60, GRM4,
PLCB2, ADCY8, ADCY6, TAS2R42, TAS1R2, TAS1R1, TRPM5 ACCN1 |
|
| Propanoate
metabolism | 31 | 0 | ACSS2 ALDH1B1,
ABAT, LOC283398, ALDH2, ACADM, ACAT2, ACAT1, LDHC, MCEE |
|
| Wnt signaling
pathway | 144 | 0 | JUN, LRP5, LRP6,
PPP3R2, SFRP2, SFRP1, PPP3CC, VANGL1, PPP3R1, FZD1 |
|
| Arrhythmogenic
right ventricular cardiomyopathy | 73 | 0 | CACNA2D1,
CACNB1, LOC100418883, CACNB2, CACNB3, CACNB4, CACNG1, ITGA9,
CACNG8, RYR2 |
|
| Axon guidance | 127 | 0 | UNC5B, PLXNB2,
PPP3R2, PPP3CC, PPP3R1, PAK4, NGEF, SEMA4C, SEMA4A, PLXNC1 |
|
| Prion diseases | 35 | 0 | NCAM2, EGR1,
NCAM1, ELK1, NOTCH1, PRKX, C6, CCL5, C5, IL1B |
|
| Cell adhesion
molecules | 125 | 0 | CDH5, JAM3,
CDH3, NLGN3, CDH4, CD80, NLGN1, CD86, CD28, CD274 |
|
| Neuroactive ligand
receptor interaction | 252 | 0 | PTGFR, PTGER2,
PTGER1, PTGER4, PTGER3, CALCRL, TACR3, PTGIR, ADRB3, ADRB2 |
|
| Olfactory
transduction | 114 | 0 | CALM2, CALM1,
OR11H4, OR52W1, OR5AU1, ADRBK2, OR2M2, OR2M7, OR2T33,
OR4F5 |
Screening for transcription factors associated with
the genes in the enriched pathways identified 123 transcription
factors associated with the genes in the CAMs pathway. Furthermore,
94 transcription factors were associated with the genes enriched in
the Wnt signaling pathway, and 87 transcription factors were
associated with the genes enriched in the PPP (Table III).
 | Table III.Counts of transcription factors for
the differentially expressed genes in the enriched pathways. |
Table III.
Counts of transcription factors for
the differentially expressed genes in the enriched pathways.
| Group | KEGG pathway | Counts |
|---|
| Antisense
miRNA-221-transfected MCF7-FR cells | PPP | 87 |
|
| Histidine
metabolism | 80 |
|
| Olfactory
transduction | 95 |
| Antisense
miRNA-222-transfected MCF7-FR cells | PPP | 87 |
|
| Taste
transduction | 76 |
|
| Propanoate
metabolism | 76 |
|
| Wnt signaling
pathway | 94 |
|
| Arrhythmogenic
right ventricular cardiomyopathy | 116 |
|
| Axon guidance | 104 |
|
| Prion diseases | 79 |
|
| Cell adhesion
molecules | 123 |
|
| Neuroactive ligand
receptor interaction | 113 |
|
| Olfactory
transduction | 95 |
Regulatory network analysis
According to the TargetScan and miRanda databases,
530 genes were targets of miRNA-221 and 488 genes were targets of
miRNA-221. Of the 530 target genes of miRNA-221, six were DEGs,
including four upregulated genes [recombination activating gene 1,
leucine-rich repeats and calponin homology domain containing 2,
methionine sulfoxide reductase B3 (MSRB3) and neurexophilin
1 (NXPH1)], in the antisense miRNA-221-transfected MCF7-FR
cells. Of the 488 target genes of miRNA-222, ten were DEGs,
including eight upregulated genes (MSRB3, NXPH1,
protocadherin (PCDH) A2, PCDH10, chromosome 9
open reading frame 135, prostaglandin E receptor 2, MAGE-like-2 and
family with sequence similarity 160 member B1), in the antisense
miRNA-222-transfected MCF7-FR cells. The miRNA-target regulatory
network is represented in Fig. 2.
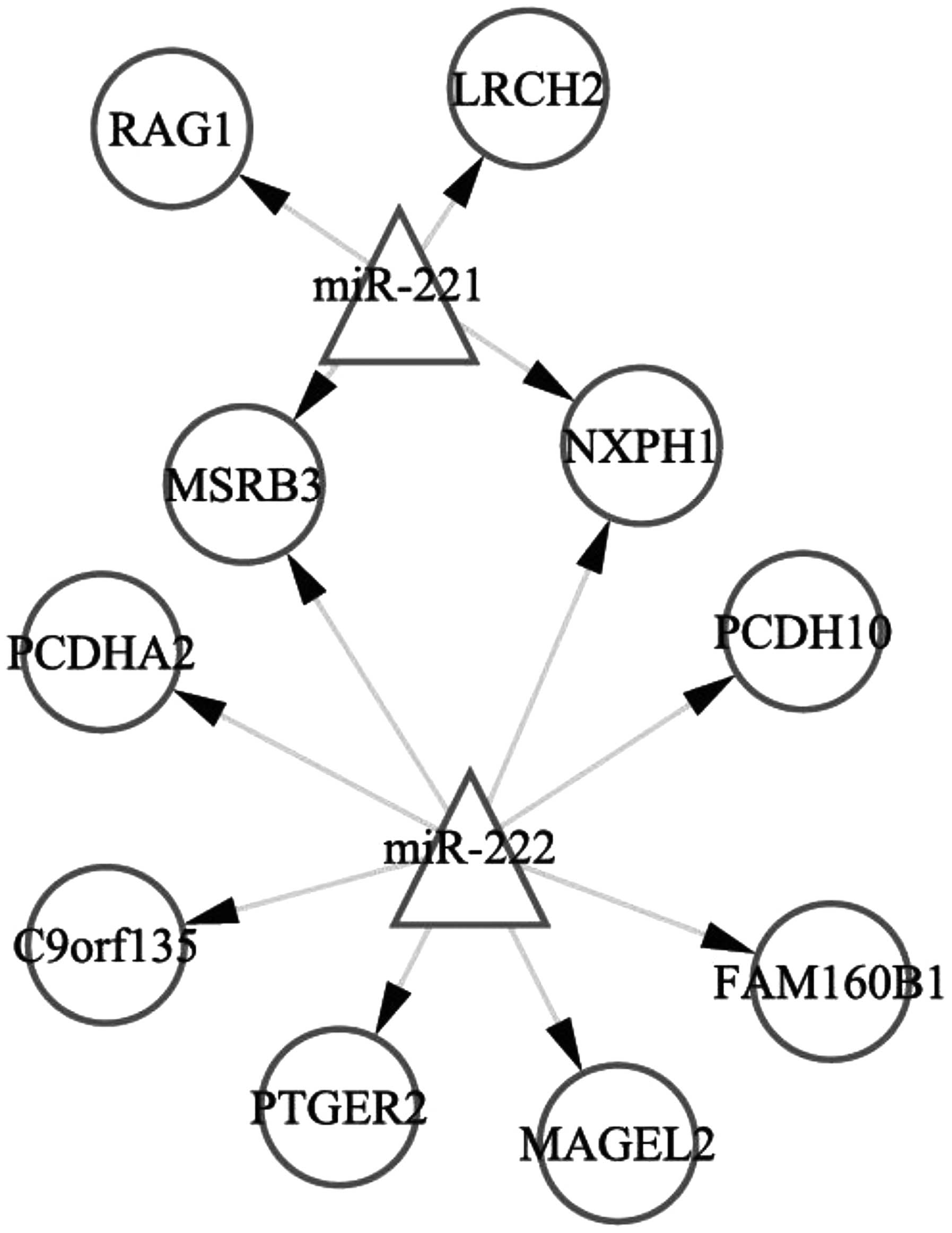 | Figure 2.miRNA (miRNA-221 and miRNA-222)-target
regulatory network. Triangles represent the miRNAs; circles
represent the differentially expressed genes in miRNA-221- and
miRNA-222-transfected MCF7-FR breast cancer cells. miRNA, microRNA;
RAG1, recombination-activating gene 1; LRCH2, leucine-rich repeats
and calponin homology domain containing 2; MSRB3, methionine
sulfoxide reductase B3; NXPH1, neurexophilin 1; PCDHA2,
protocadherin α-2; PCDH10, protocadherin-10; C9orf135, chromosome 9
open reading frame 135; PTGER2, prostaglandin E receptor 2; MAGEL2,
MAGE family member L2; FAM160B1, family with sequence similarity
160 member B1. |
Discussion
In the present study, 492 and 404 DEGs were
identified in the antisense miRNA-221-transfected MCF7-FR cells and
the antisense miRNA-222-transfected MCF7-FR cells, respectively, as
compared with the negative control. GSEA revealed that the PPP was
significantly enriched in the antisense miRNA-221-transfected and
antisense miRNA-222-transfected MCF7-FR cells. Furthermore, 87
transcription factors were identified for the genes enriched in the
PPP, which suggested that the PPP was significantly regulated in
these cells. The PPP produces two substrates, ribose 5-phosphate
and nicotinamide adenine dinucleotide phosphate, which are
necessary for the division of cells and serve as buffers to prevent
reactive oxygen species-induced cell death and apoptosis (17). Alterations in the PPP activity have
been reported to occur during cancer development and progression
(18). In addition, an increase in
the levels of various PPP metabolites in the breast epithelia,
including sedoheptulose 7-phosphate and hexose phosphate
intermediates, has been reported to occur during the transition
from normal breast epithelial cells to transformed cells, as well
as during the transition from non-metastatic to metastatic tumors
(19,20).
In the present study, the Wnt signaling pathway was
significantly enriched in the antisense miRNA-222-transfected
MCF7-FR cells compared with the normal control-transfected cells. A
total of 94 transcription factors were associated with the genes
enriched in the Wnt signaling pathway, which suggested that this
pathway was highly regulated in the miRNA-222-transfected MCF7-FR
cells. A previous study reported that the activation of the Wnt
signaling pathway could lead to the metastasis of breast cancer
(21). In addition, the blockage of
Wnt signaling has been demonstrated to inhibit cell proliferation
and migration, and to induce apoptosis in triple-negative breast
cancer cells (22).
In the present study, the CAMs pathway was
significantly enriched in the antisense miRNA-222-transfected
MCF7-FR cells compared with the normal control-transfected cells. A
total of 123 transcription factors were associated with the genes
enriched in this pathway. CAMs are membrane receptors that mediate
cell-cell and cell-matrix interactions, and have an essential role
in transducing intracellular signals responsible for adhesion,
migration, invasion, angiogenesis and organ-specific metastasis
(23). Adhesion molecules, including
E-cadherin and carcinoembryonic antigen, have been associated with
the process of metastasis in breast cancer cells (24). Taken together, these results suggested
that the PPP, Wnt signaling pathway and CAMs pathway may be
associated with the resistance of breast cancer to fulvestrant.
In the miRNA-target regulatory network, miR-222 was
observed to target PCDH10. PCDH10 is a member of the
mammalian cadherin superfamily, which has key roles in cell
migration and calcium-dependent, cadherin-mediated homophilic
cell-cell interactions (25). A
previous study identified PCDH10 as a candidate tumor
suppressor in nasopharyngeal, esophageal and various other
carcinomas, in which it was associated with frequent methylation
(26). As a result, PCDH10
targeted by miR-222 could be associated with the resistance of
breast cancer to fulvestrant.
In conclusion, the results of the present study
suggested that the PPP, Wnt signaling pathway and CAMs KEGG
pathway, as well as PCDH10, may be associated with the
development of fulvestrant resistance in patients with breast
cancer. However, further studies are required to elucidate the
underlying mechanisms.
Acknowledgements
The present study was supported by the Health Bureau
Science and Technology Foundation of Tianjin (Tianjin, China; grant
no. 2012KZ063) and the National Natural Science Foundation of China
(Beijing, China; grant nos. 81302082, 81272685, 31301151 and
81172355).
References
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ariazi EA, Ariazi JL, Cordera F and Jordan
VC: Estrogen receptors as therapeutic targets in breast cancer.
Curr Top Med Chem. 6:181–202. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Howell A: Fulvestrant (‘Faslodex’):
Current and future role in breast cancer management. Crit Rev Oncol
Hematol. 57:265–273. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Baumgarten SC and Frasor J: Minireview:
Inflammation: An instigator of more aggressive estrogen receptor
(ER) positive breast cancers. Mol Endocrinol. 26:360–371. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Xin F, Li M, Balch C, Thomson M, Fan MY,
Liu Y, Hammond SM, Kim S and Nephew KP: Computational analysis of
microRNA profiles and their target genes suggests significant
involvement in breast cancer antiestrogen resistance.
Bioinformatics. 25:430–434. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Rao X, Di Leva G, Li M, Fang F, Devlin C,
Hartman-Frey C, Burow ME, Lvan M, Croce CM and Nephew KP:
MicroRNA-221/222 confers breast cancer fulvestrant resistance by
regulating multiple signaling pathways. Oncogene. 30:1082–1097.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Perey L, Paridaens R, Hawle H, Zaman K,
Nolé F, Wildiers H, Fiche M, Dietrich D, Clément P, Köberle D, et
al: Clinical benefit of fulvestrant in postmenopausal women with
advanced breast cancer and primary or acquired resistance to
aromatase inhibitors: Final results of phase II Swiss Group for
Clinical Cancer Research Trial (SAKK 21/00). Ann Oncol. 18:64–69.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Tangkeangsirisin W and Serrero G: GP88
(Progranulin) confers fulvestrant (Faslodex, ICI 182,780)
resistance to human breast cancer cells. Adv Breast Cancer Res.
3:68–78. 2014. View Article : Google Scholar
|
|
9
|
Kirkegaard T, Yde CW, Kveiborg M and
Lykkesfeldt AE: The broad-spectrum metalloproteinase inhibitor
BB-94 inhibits growth, HER3 and Erk activation in
fulvestrant-resistant breast cancer cell lines. Int J Oncol.
45:393–400. 2014.PubMed/NCBI
|
|
10
|
Afsari B, Geman D and Fertig EJ: Learning
dysregulated pathways in cancers from differential variability
analysis. Cancer Inform. 13(Suppl 5): S61–S67. 2014.
|
|
11
|
Troyanskaya O, Cantor M, Sherlock G, Brown
P, Hastie T, Tibshirani R, Botstein D and Altman RB: Missing value
estimation methods for DNA microarrays. Bioinformatics. 17:520–525.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Székely GJ and Rizzo ML: Hierarchical
clustering via joint between-within distances: Extending Ward's
minimum variance method. J Classif. 22:151–183. 2005. View Article : Google Scholar
|
|
13
|
Deza E and Deza MM: Encyclopedia of
distances. 3rd. Springer-Verlag; Berlin: 2009, View Article : Google Scholar
|
|
14
|
Shi J and Walker MG: Gene Set Enrichment
Analysis (GSEA) for Interpreting Gene Expression Profiles. Curr
Bioinform. 2:133–137. 2007. View Article : Google Scholar
|
|
15
|
Gotea V and Ovcharenko I: DiRE:
Identifying distant regulatory elements of co-expressed genes.
Nucleic Acids Res. 36:(Web Server Issue). W133–W139. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Smoot ME, Ono K, Ruscheinski J, Wang PL
and Ldeker T: Cytoscape 2.8: New features for data integration and
network visualization. Bioinformatics. 27:431–432. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Tian WN, Braunstein LD, Apse K, Pang J,
Rose M, Tian X and Stanton RC: Importance of glucose-6-phosphate
dehydrogenase activity in cell death. Am J Physiol.
276:C1121–C1131. 1999.PubMed/NCBI
|
|
18
|
Riganti C, Gazzano E, Polimeni M, Aldieri
E and Ghigo D: The pentose phosphate pathway: An antioxidant
defense and a crossroad in tumor cell fate. Free Radic Biol Med.
53:421–436. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Richardson AD, Yang C, Osterman A and
Smith JW: Central carbon metabolism in the progression of mammary
carcinoma. Breast Cancer Res Treat. 110:297–307. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lu X, Bennet B, Mu E, Rabinowitz J and
Kang Y: Metabolomic changes accompanying transformation and
acquisition of metastatic potential in a syngeneic mouse mammary
tumor model. J Biol Chem. 285:9317–9321. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Cai J, Guan H, Fang L, Yang Y, Zhu X, Yuan
J, Wu J and Li M: MicroRNA-374a activates Wnt/β-catenin signaling
to promote breast cancer metastasis. J Clin Invest. 123:566–579.
2013.PubMed/NCBI
|
|
22
|
Bilir B, Kucuk O and Moreno CS: Wnt
signaling blockage inhibits cell proliferation and migration, and
induces apoptosis in triple-negative breast cancer cells. J Transl
Med. 11:2802013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Li DM and Feng YM: Signaling mechanism of
cell adhesion molecules in breast cancer metastasis: Potential
therapeutic targets. Breast Cancer Res Treat. 128:7–21. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Saadatmand S, de Kruijf EM, Sajet A,
Dekker-Ensink NG, van Nes JG, Putter H, Smit VT, van de Velde CJ,
Liefers GJ and Kuppen JK: Expression of cell adhesion molecules and
prognosis in breast cancer. Br J Surg. 100:252–260. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Pines J, Toldo L and Lafont F: Cell to
cell contact and extracellular matrix. Curr Opin Cell Biol.
10:5611998. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ying J, Li H, Seng TJ, Langford C,
Srivastava G, Tsao SW, Putti T, Murray P, Chan AT and Tao Q:
Functional epigenetics identifies a protocadherin PCDH10 as a
candidate tumor suppressor for nasopharyngeal, esophageal and
multiple other carcinomas with frequent methylation. Oncogene.
25:1070–1080. 2006. View Article : Google Scholar : PubMed/NCBI
|