Introduction
Chronic myeloid leukemia (CML) refers to a group of
neoplasias that are defined by a unique genetic aberration, the
breakpoint cluster region-Abelson murine leukemia viral oncogene
homolog 1 (BCR-ABL1) fusion gene (1).
CML is also reognized as one of the best examples for molecular
targeted therapy (2). However,
numerous aspects of the pathogenesis of CML have not been
elucidated thus far, including the mechanisms of blastic crises,
the causes of genetic instabilities such as the inactivation of
tumor suppressor genes, and the oncogenic signaling pathways
downstream of the BCR-ABL1 fusion gene product (3).
In recent years, microvesicles (MVs) in the leukemia
microenvironment have been drawn researchers' attention (4,5). MVs, also
known as exosomes or microparticles, are vesicles comprised of
various cell organelles of diameters ranging from 30 to 1,000 nm
(6). The formation of MVs can happen
on the cellular membrane surface or the endosome (7). The functions of MVs are closely
associated with the contents in the vesicle (8). Despite their small size, MVs are
enriched in bioactive molecule, including growth factors and their
receptors, proteinases, adhesion molecules, signal molecules, DNA,
messenger (m)RNA and microRNA (also known as miR) (4). These bioactive molecules collectively
function as the signaling complex (9–12). It has
been reported that MVs released from CML cells are important in
angiogenesis (13,14) and immunosuppression (15). The mechanisms of these MVs' roles have
not been yet elucidated. Previous studies revealed that genetic
exchanges of microRNAs between cells can be accomplished through
MVs (16–18). MVs can fuse with the cellular membrane
and transfer all the aforementioned bioactive molecules to receptor
cells (17), thus being important in
cell-to-cell communication.
MicroRNAs are non-coding, single-stranded RNAs of
21–25 nucleotides, which have recently been implicated in the
regulation of a number of biological processes, including
development, differentiation, apoptosis, proliferation and
hematopoiesis (19). MicroRNAs
regulate gene expression by promoting the degradation of their
target mRNA or repressing its translation (19). MicroRNA expression is tissue-specific,
and has been demonstrated to be altered in a number of human
cancers (20,21). In CML, abnormal expression of several
microRNAs has been described, including miR-15a, miR-16, miR-142,
miR-155, miR-181, miR-221, let7a and the polycistronic miR-17-92
cluster (22–24).
Notably, a connection between MVs and microRNA has
been recently proposed (25).
Specific miRNAs regulate hematopoietic cell differentiation and
development (26). Previous studies
have indicated that microRNA packaged within MVs may be transported
extracellularly (25,27–30). It is
possible that these microRNAs in MVs reflect the miRNA signature of
the parental tumor (31). Employing
MVs to transfer genetic material would be an efficient transfer
method between cells, and MVs containing microRNAs would enable
intercellular and inter-organ communication in the body.
The present study explored the hypothesis that
microRNAs are contained within MVs derived from the CML cell line
K562. The microRNA expression profile of MVs from K562 cells and
from normal human volunteers' peripheral blood cells was examined,
and the microRNA expression in K562 cells-derived MVs was compared
with that in K562 cells, and their roles in leukemia were analyzed.
The objectives of the present study were to determine whether the
microRNAs contained within leukemia-cell-derived MVs mirrored those
of leukemia cells and thus, could be used diagnostically and
therapeutically.
Materials and methods
Cell line and control samples
The K562 cells (kindly provided by Dr.Jingqiong Hu,
Tongji Medical College, Huazhong University of Science and
Technology, Wuhan, China) were grown in RPMI 1640 medium (Gibco;
Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with
10% MV-free (by ultrafiltration) fetal bovine serum (Gibco; Thermo
Fisher Scientific, Inc.), 25 mM
4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid, 24 mM sodium
bicarbonate, 2 mM L-glutamine, 100 IU/ml penicillin and 100 IU/ml
streptomycin in a humidified 5% CO2 atmosphere at 37°C.
Cell viability was evaluated by trypan blue exclusion assay, and
all cultures utilized were 95% viable. A method was established
using ultrafiltration to remove MVs from fetal bovein serum.
Initially, debris in fetal bovein serum was removed by 0.22 µm
filter (EMD Millipore, Billerica, MA, USA). Subsequently, the
supernatant was purified by stirring ultrafiltration with a 100,000
molecular weight cut off ultrafiltration membrane (EMD Millipore)
to remove MVs.
Peripheral blood was collected in
ethylenediaminetetraacetic acid-coated tubes from healthy donors at
Union Hospital (Tongji Medical College, Huazhong University of
Science and Technology) between August and October 2012. The
criteria for volunteers selection consisted of no recent illnesses
or treatments for a chronic medical condition. No medical history
was obtained from the donors. The collection of blood occurred
between the morning and early afternoon. The average age for the
female and male donors was 35.79 and 31.81 years, respectively.
Written informed consent was obtained from all volunteers and this
study was approved by the ethics committee of Tongji Medical
College, Huazhong University of Science and Technology.
Isolation of MVs from cell culture
supernatants
Supernatants were collected from cell culture after
24 h. Cells were centrifuged at 300 × g for 30 min, and the
supernatant was used for MVs preparation. Cell debris was removed
by centrifugation at 2,500 × g for 30 min. MVs were purified by
ultracentrifugation at 16,000 × g for 120 min, and subsequently
washed with sterile filtered phosphate-buffered saline (PBS). The
samples were resuspended in radioimmunoprecipitation assay (RIPA)
buffer supplemented with a protease inhibitor cocktail (Agilent
Technologies, Inc., Santa Clara, CA, USA) and stored at −80°C. All
centrifugations were accomplished at 4°C. MVs quantity was
determined with the Pierce BCA Protein Assay kit (Thermo Fisher
Scientific, Inc., Waltham, MA, USA).
Isolation of MVs from normal human
volunteers' peripheral blood cells
Peripheral blood was centrifuged at 400 × g for 30
min, and the plasma was used for MVs preparation. Cell debris and
platelets were removed by centrifugation at 2,500 g for 20 min
twice. MVs were purified by ultracentrifugation at 16,000 × g for
120 min, and subsequently washed with sterile filtered PBS
(32). All centrifugations were
accomplished at 4°C. MV pellets were resuspended in RIPA buffer
supplemented with a protease inhibitor cocktail (Agilent
Technologies, Inc.) and stored at −80°C. MVs quantity was
determined with the Pierce BCA Protein Assay kit (Thermo Fisher
Scientific, Inc.).
Transmission electron microscopy
For transmission electron microscopy, the pelleted
MVs were fixed in 2.5% (w/v) glutaraldehyde in PBS, dehydrated and
embedded in Epon (SPI Supplies, Inc., West Chester, PA, USA).
Ultrathin sections (65-nm) were cut and stained with uranyl acetate
(SPI Supplies, Inc.) and Reynold's lead citrate (SPI Supplies,
Inc.). The sections were examined in a Tecnai G2 12
transmission electron microscope (FEI, Hillsboro, OR, USA).
RNA extraction and purification
Total RNA was isolated from K562 cells and MVs using
the mirVana microRNA Isolation kit according to the manufacturer's
protocol (Ambion; Thermo Fisher Scientific, Inc.). The quality,
yield and size of the microRNA fractions were analyzed using a 2100
Bioanalyzer (Agilent Technologies, Inc.). For RNA isolated from
mononuclear cells, only an RNA integrity number (RIN) ≥7 was used,
along with its matched plasma sample for profiling. Since the
intact 18 and 28s ribosomal RNA were variable in the MVs, the RIN
was not a constraint for these samples, although a RIN between 6.5
and 6.8 was observed.
MicroRNA profiling
RNA from K562 cells, and MVs both from K562 cells
and normal human volunteers' peripheral blood cells were used for
microRNA microarray. Human microRNA microarrays from Agilent
Technologies, Inc., which contain probes for 888 human microRNAs
from the miRBase v14.0 (mirbase.org/pub/mirbase/14.0/), were used in the
study. In total, 100 ng of total RNA extracted from serum was used
as inputs for sample labeling and hybridization preparation,
following the manufacturer's protocol.
The microarray image information was converted into
spot intensity values using Scan Control Software version 7.0
(Agilent Technologies, Inc.). The signal upon background
subtraction was exported directly into the GeneSpring GX 11.0
software (Agilent Technologies, Inc.) for quartile normalization
and further analysis.
Validation of microarray data
For testing of candidate microRNAs acquired on
microarrays, reverse transcription-quantitative polymerase chain
reaction (RT-qPCR) was performed using miScript SYBR Green PCR kit
(Qiagen GmbH, Hilden, Germany). The assays were performed on three
samples for six candidates (miR-494, miR-483-5p, miR-26a, miR-223,
miR-21 and miR-22).
Statistical analysis
Data were analyzed using the statistical software
package SAS v3.0. (Shanghai Biotechnologies Corporation, Shanghai,
China). The statistical significance was calculated by the
Student's t test. P≤0.05 was considered to indicate a
statistically significant difference. Three-way Venn diagrams were
used to indicate the number of significant microRNAs [P<0.05,
fold-change (FC)>2 and FC<0.5] identified by the Student's
t test (K562-MVs vs. control MVs).
Pathway analysis and prediction
The human TargetScan Release 5.1 (http://www.targetscan.org) was used for prediction of
microRNA targets. In addition, potential target gene-associated
pathways were analyzed using TargetScan Release 5.1 based on the
Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway database
(http://www.genome.jp/kegg/). The
enrichment P-value of the target genes involved in every pathway
was calculated, and the regulatory interactions between genes and
microRNAs were integrated.
Results
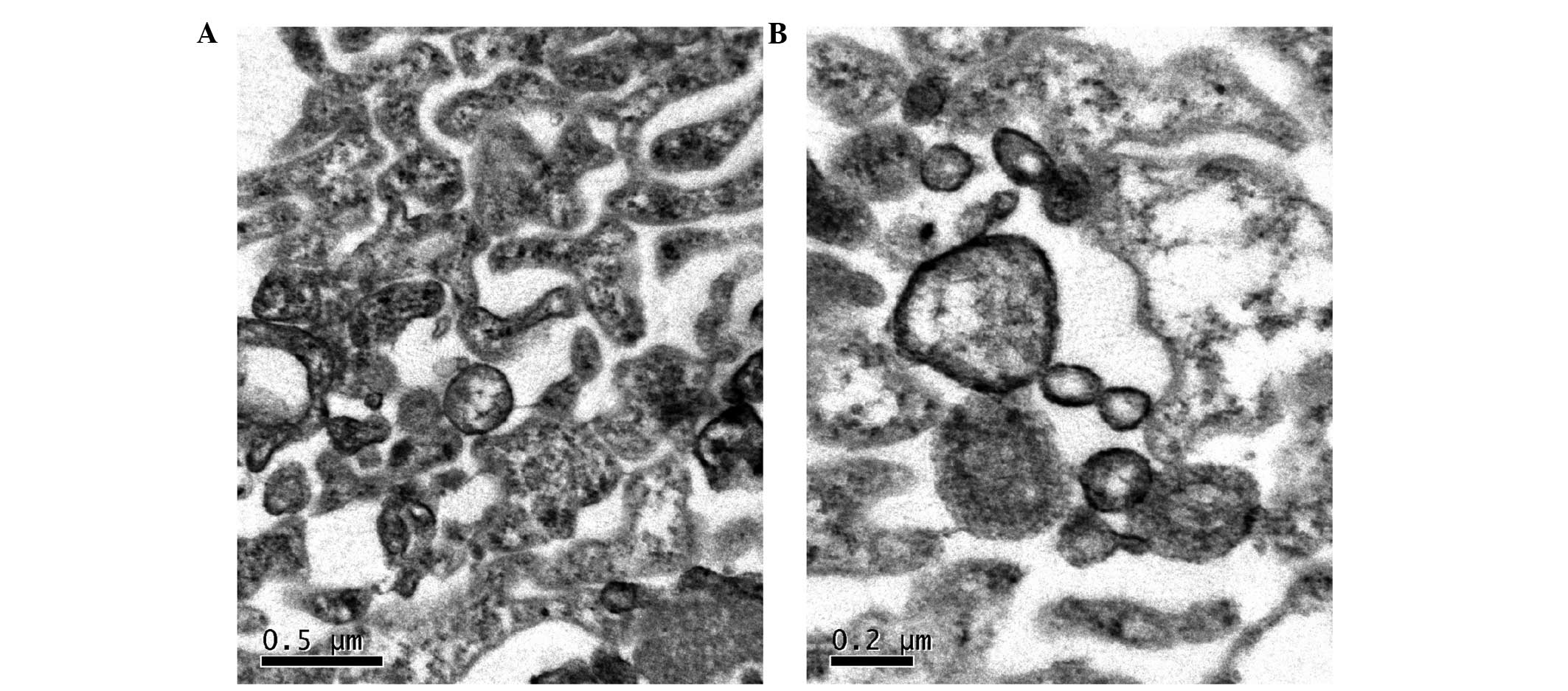
Transmission electron microscopy
MVs derived from K562 cells and normal human
volunteers' peripheral blood cells were collected and observed
under a transmission electron microscope, which revealed vesicular
structures characteristic of MVs (Fig. 1A
and B).
MicroRNA expression profile in
K562-derived MVs
A microarray containing probes for 888 human
microRNAs was initially used to screen the significant differential
expression levels of microRNAs between K562-MVs and control groups
(Table I). The filtered and
normalized data were subjected to hierarchical cluster analysis
comparing the microRNA expression profile of K562-MVs and control
group samples. Fig. 2 illustrates the
hierarchical clustering of the differentially expressed microRNAs
in the pairwise comparison of the two samples. There were seven
microRNAs, including miR-494, miR-1275, miR-484-5p, miR-1308-v15.0,
miR-575, miR-1268 and miR-125a-3p, with significantly higher
expression levels in the K562-MV group than in the healthy group
(FC=7.18–96.49; P<0.05; Table I).
By contrast, miR-151-3p, miR-1974-v14.0, miR-26a, miR-24, miR-22,
miR-93, miR-223, miR-23b, miR-103, miR-361-5p, miR-21, miR-126*,
miR-107, miR-27b, miR-27a and miR-185 displayed a significantly
lower expression level in the K562-MV group than in the healthy
group (FC<0.5; P<0.05; Table
I).
 | Table I.Expression signatures of dysregulated
miRNAs from K562-derived MVs. |
Table I.
Expression signatures of dysregulated
miRNAs from K562-derived MVs.
| A, Upregulated
microRNAs in K562-derived MVs |
|---|
|
|---|
| Name | P-value | Fold-change |
|---|
| miR-151-3p | 0.009770351 | 0.0134710 |
| miR-1974_v14.0 | 0.003311413 | 0.0146479 |
| miR-26a | 0.003024278 | 0.0146984 |
| miR-223 | 0.049822803 | 0.0147095 |
| miR-23b | 0.024271710 | 0.0390509 |
| miR-103 | 0.023439787 | 0.0406894 |
| miR-24 | 0.003045386 | 0.0496698 |
| miR-361-5p | 0.036743575 | 0.0531059 |
| miR-21 | 0.024356252 | 0.0796379 |
| miR-22 | 0.007922998 | 0.0820633 |
| miR-126* | 0.022427225 | 0.0843179 |
| miR-107 | 0.048353604 | 0.1005024 |
| miR-27b | 0.033929142 | 0.1036153 |
| miR-93 | 0.000452252 | 0.1485404 |
| miR-27a | 0.021547485 | 0.1844731 |
| miR-185 | 0.039805662 | 0.4350724 |
|
| B, Downregulated
microRNAs in K562-derived MVs |
|
| Name | P-value | Fold-change |
|
| miR-494 | 0.033709016 | 7.1837779 |
| miR-1275 | 0.040862084 | 11.0616334 |
| miR-483-5p | 0.030694947 | 12.8443913 |
| miR-1308_v15.0 | 0.015232616 | 26.8287621 |
| miR-575 | 0.015022667 | 46.2936787 |
| miR-1268 | 0.015112414 | 59.5764594 |
| miR-125a-3p | 0.027056158 | 96.4917155 |
Comparison of K562-MVs' microRNA
expression with that of parental cells
The presence and levels of specific microRNAs from
both K562-MVs and their parental cells were determined using
microarray analysis probing for 888 human microRNAs. The microRNA
profiles of K562 cells confirmed the alterations previously
reported (33). Furthermore, the
results demonstrated that 30 microRNAs were above the normalized
threshold in the 888 microRNAs, which was calculated based on the
95 percentile of the negative control probe signal in both normal
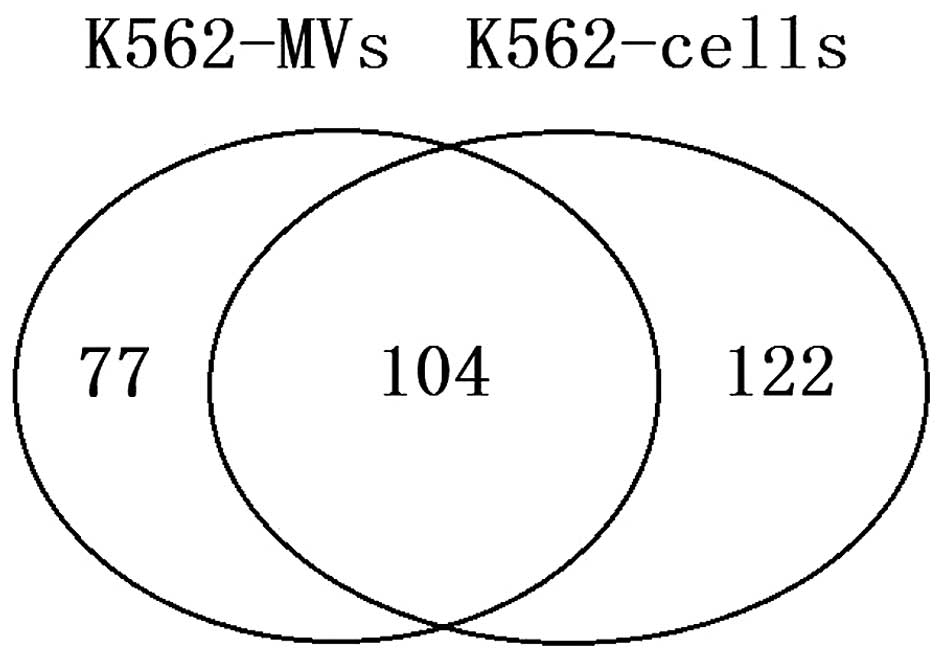
cells and MVs (Fig. 3 and Table II). Of the 303 positive microRNAs,
104 were not significantly different between MVs and their parental
K562 cells. By comparison, 77 microRNAs were present at elevated
levels within MVs, while 122 were present at a higher proportion in
the parental cells. This observation may suggest that the
compartmentalization of microRNAs from cells into MVs is an active
(selective) process, at least for certain microRNAs.
 | Table II.Expression levels of microRNAs in
K562 cells-derived MVs compared with those of microRNAs isolated
from K562 cells. |
Table II.
Expression levels of microRNAs in
K562 cells-derived MVs compared with those of microRNAs isolated
from K562 cells.
| Elevated in
cells | Equal in cells and
MVs | Elevated in
MVs |
|---|
| miR-142-5p,
miR-374a, miR-590-5p, | let-7a, let-7b,
let-7c, let-7e, | miR-630,
miR-671-5p, |
| miR-101, miR-29b,
miR-20a*, miR-377, | let-7f,
hsa-let-7f-1*, let-7g, | miR-874,
miR-188-5p, |
| miR-144, miR-381,
miR-374b, miR-424, | let-7i, miR-103,
miR-106b, | miR-483-5p,
miR-513a-5p, |
| miR-21*,
miR-140-5p, miR-301a, | miR-107,
miR-1202, | miR-1224-5p,
miR-765, |
| miR-19b-1*,
miR-29c, miR-342-3p, | miR-125a-3p,
miR-125a-5p, | miR-654-5p,
miR-135a*, |
| miR-486-3p,
miR-135b, miR-10a, | miR-125b,
miR-126, | miR-1226*,
miR-892b, |
| miR-154*, miR-301b,
miR-127-3p, | miR-1268,
miR-1275, | miR-1183,
miR-2276, |
| miR-379, miR-487b,
miR-140-3p, | miR-1305,
miR-130b, | miR-584,
miR-371-5p, |
| miR-136,
miR-654-3p, miR-299-3p, | miR-142-3p,
miR-151-3p, | miR-500a,
miR-150*, |
| miR-331-3p,
miR-431*, miR-33a, | miR-151-5p,
miR-15a, | miR-422a,
miR-520b, |
| miR-410, miR-218,
miR-660, miR-148b, | miR-15b, miR-16,
miR-17, | miR-155,
miR-501-5p, |
| miR-212, miR-223*,
miR-100, miR-148a, | miR-17*,
miR-185, | miR-921,
miR-628-3p, |
| miR-409-3p,
miR-30e*, miR-495, | miR-19a,
miR-19b, | miR-1287,
miR-887, |
| miR-99b, miR-192,
miR-337-5p, miR-186, | miR-20a, miR-20b,
miR-21, | miR-1471,
miR-520e, |
| miR-32, miR-493*,
miR-299-5p, | miR-210,
miR-22, | miR-149*,
miR-500a*, |
| miR-376b, miR-183
and miR-324-5p | miR-223,
miR-224, | miR-718,
miR-1181, |
|
| miR-23a, miR-23b,
miR-24, | miR-513c,
miR-542-5p, |
|
| miR-25, miR-26a,
miR-26b, | miR-564,
miR-1299, |
|
| miR-27a, miR-27b,
miR-29a, | miR-662,
miR-622, |
|
| miR-30b, miR-30c,
miR-30e, | miR-490-5p,
miR-557, |
|
| miR-494,
miR-575, | miR-877,
miR-602, |
|
| miR-638,
miR-7, | miR-610,
miR-760, |
|
| miR-92a and
miR-93 | miR-125b-1*,
miR-202, |
|
|
| miR-502-3p,
miR-99b*, |
|
|
| miR-663,
miR-583 |
|
|
| and miR-617 |
Validation of microarray data
Two upregulated (miR-494 and miR-484-5p) and four
downregulated (miR-26a, miR-22, miR-223 and miR-21) microRNAs of
K562-MVs (in terms of their expression levels compared with those
in the control) were selected for microarray data validation via
RT-qPCR, and the results correlated well with the findings of
microarray analysis.
Putative microRNA target genes
analysis and functional annotation
To further study the functions of the aberrantly
expressed microRNAs in MVs and in their parental cells, a predicted
target analysis for these microRNAs was performed. The data
revealed that 23 microRNAs exhibited altered expression in
K562-derived MVs compared with control MVs. These microRNAs
targeted 1,354 regulatory genes, which affect cellular apoptosis,
proliferation and molecular signaling pathways. Notably, 43
oncogenes and tumor suppressor genes were identified among these
aberrant microRNAs detected in MVs derived from K562 cells
(Table III). In these dysregulated
microRNAs, 4 microRNAs targeted 2 oncogenes of the B-cell lymphoma
family, while 12 microRNAs targeted 16 oncogenes of the rat sarcoma
viral oncogene homolog (RAS) family. In addition, 24 microRNAs
targeted 22 housekeeping genes in the p53 signaling pathway
(Table IV). p53 is a tumor
suppressor protein that regulates the expression of a wide variety
of genes (34). It has been reported
that the p53 signaling pathway has a key role in the induction of
apoptosis of CML cells (35). With
the exception of breast cancer 1, which was targeted by
miR-125a-3p, and insulin-like growth factor 1, which was targeted
by miR-1275 and miR-1207-5p, 16 of the above housekeeping genes
were targeted by the downregulated microRNAs.
 | Table III.Tumor related genes targeted by
miRNAs from K562-derived MVs. |
Table III.
Tumor related genes targeted by
miRNAs from K562-derived MVs.
| A, Oncogenes
targeted by microRNAs in K562-derived MVs |
|---|
|
|---|
| Oncogene | microRNA
species | Oncogene | microRNA
species |
|---|
| BCL3 |
miR-27b|miR-27a | RAB4B | miR-24 |
| BCL7A |
miR-1202|miR-21 | RAB5A | miR-494 |
| CASC4 | miR-575 | RAB8B |
miR-23a|miR-23b |
| CBL | miR-425 | RAP1A | miR-24 |
| ERBB3 | miR-22|miR-24 | RAP2C | miR-93|miR-24 |
| ERBB4 | miR-193b|
miR-23a|miR-23b | RAN | miR-134 |
| ETS1 | miR-1202 | RASL11B | miR-93 |
| FYN | miR-125a-3p | RASSF2 | miR-93 |
| MCL1 | miR-93 | RASD1 | miR-93 |
| MLF2 | miR-93 | RBBP4 | miR-24 |
| MTUS1 | miR-361-5p | RHOU |
miR-26a|miR-26b |
| MYCT1 |
miR-23a|miR-23b | RHOC | miR-93 |
| RAB11A | miR-21 | RRAS2 | miR-23a,
miR-23b|miR-223 |
| RAB21 |
miR-26a|miR-26b | SKI | miR-21 |
| RAB35 | miR-185 | STMN1 | miR-193b |
| RAB39B |
miR-23a|miR-23b | TET3 |
miR-26a|miR-26b |
|
| B, Tumor
suppressors targeted by microRNAs in K562-derived MVs |
|
| Tumor
suppressor | microRNA
species | Tumor
suppressor | microRNA
species |
|
| BAK1 |
miR-26a|miR-26b|miR-27b|miR-27a | NF1 |
miR-103,miR-107|miR-128| miR-125a-3p |
| BCR |
miR-26a|miR-26b | PHF6 | miR-26a |
| BRCA1 | miR-125a-3p | PTEN |
miR-22|miR-494|miR-29a miR-29b |
| BRMS1L | miR-93 | RGS4 |
miR-26a|miR-26b |
| BTG3 | miR-93 | TP53INP1 | miR-22 |
| FOXO1 | miR-223 | TUSC2 |
miR-23a|miR-93|miR-23b |
| IKZF4 | miR-575 |
|
|
 | Table IV.Housekeeping genes in the p53
signaling pathway that are targeted by microRNAs in K562-derived
microvesicles. |
Table IV.
Housekeeping genes in the p53
signaling pathway that are targeted by microRNAs in K562-derived
microvesicles.
| MicroRNAs | Genes | Role in the p53
pathway |
|---|
| miR-185 | BAI1 | Negative regulation
of cell proliferation, inhibition of angiogenesis and
metastasis |
|
miR-26a|miR-26b|miR-27b|miR-27a | BAK1 | Apoptosis |
|
miR-26a|miR-26b | BID | Induction of
apoptosis |
| miR-125a-3p | BRCA1 | Apoptosis, DNA
repair and cell proliferation |
| miR-93 | CASP2 | Anti-apoptosis |
| miR-93 | CCNG2 | Cell cycle
checkpoint |
|
miR-23a|miR-23b | CCNH | Cell cycle
genes |
|
miR-27b|miR-27a | CHEK2 | Cell cycle
arrest |
| miR-22 | CYR61 | Cell
proliferation |
|
miR-1275|miR-1207-5p | IGF1 | Anti-apoptosis |
|
miR-23a|hsa-let-7f|hsa-let-7e|hsa-let-7c | FAS | Apoptosis |
|
hsa-let-7b|hsa-let-7a|miR-23b|hsa-let-7g
miR-21 | MSH2 | DNA repair genes
and negative regulation of the cell cycle |
|
miR-125a-3p|miR-103| miR-107|miR-128 | NF1 | Negative regulation
of cell proliferation and negative regulation of the cell
cycle |
|
miR-27b|miR-128|miR-27a | PHB | Associated with
cell growth, proliferation and differentiation |
|
miR-103|miR-107 | RAI14 | Apoptosis |
|
miR-27b|miR-27a | SESN2 | Cell cycle
arrest |
| miR-223 | SNCA | Anti-apoptosis |
| miR-93 | MCL1 | Anti-apoptosis |
| miR-24 | TSHR | Positive regulation
of cell proliferation |
| miR-24 | PIGS | Apoptosis |
A predicted target analysis was also performed for
the 104 microRNAs co-expressed in MVs and in their parental K562
cells. In total, 97 microRNAs targeted 3,743 regulatory genes. Data
regarding miR-17*, miR-191*, miR-1914*, miR-1974_v14.0,
miR-1977_v14.0, miR-1979_v15.0 and miR-33b* were not available in
TargetScan 5.1.
Gene ontology (GO) analysis revealed that the
high-enrichment GOs targeted by the microRNAs co-expressed in MVs
and the corresponding K562 cells were involved in various
processes, including cell communication, signal transduction, RNA
metabolism, transcription and cell differentiation. The pathway
analysis revealed that there were 13 different pathways
corresponding to the target genes, 12 of which were enriched
(P<0.01) (Table V).
 | Table V.Target gene-associated pathways. |
Table V.
Target gene-associated pathways.
| Term | Number | q-value | Enrichment test
P-value |
|---|
| Cell cycle | 3 | 0.1089 | 0.9065 |
| Notch signaling
pathway | 8 | 0.0014 | 0.0053 |
| Chronic myeloid
leukemia | 18 | 0.0000 | 0.0000 |
| VEGF signaling
pathway | 18 | 0.0000 | 0.0000 |
| mTOR signaling
pathway | 23 | 0.0000 | 0.0000 |
| TGFβ signaling
pathway | 23 | 0.0000 | 0.0000 |
| p53 signaling
pathway | 24 | 0.0000 | 0.0000 |
| ErbB signaling
pathway | 26 | 0.0000 | 0.0000 |
| Jak-STAT signaling
pathway | 26 | 0.0000 | 0.0000 |
| Wnt signaling
pathway | 41 | 0.0000 | 0.0000 |
| Insulin signaling
pathway | 46 | 0.0000 | 0.0000 |
| Focal adhesion | 61 | 0.0000 | 0.0000 |
| MAPK signaling
pathway | 90 | 0.0000 | 0.0000 |
Mitogen-activated protein kinase
(MAPK) signaling pathway is regulated by microRNAs in K562-MVs and
K562 cells
To elucidate the mechanisms by which microRNAs
co-expressed in MVs and parental K562 cells regulated
leukemogenesis, a bioinformatic approach was undertaken to identify
signaling pathways and target genes regulated by these microRNAs.
Using SAS v3.0, which is designed to integrate microRNA target
genes into signaling pathways, mRNAs involved in the MAPK signaling
pathway, focal adhesion pathway, insulin signaling pathway and Wnt
signaling pathway were observed to be significantly enriched in
these microRNA target genes. These target genes were analyzed, and
it was noticed that 34 microRNAs targeted 44 genes of the MAKP
pathway, suggesting that the MAPK signaling pathway may play a
significant role in MVs and their parental K562 cells.
The microRNA-gene interaction networks are
represented in Fig. 4. As indicated
in Fig. 4, the miR-30 family (with
the exception of miR-30a and miR-30d) contained the most targeted
mRNAs, with degrees ranging from 6 to 9, while miR-17, miR-16 and
miR-15a/b occupied an important position in the MAPK pathway. These
microRNAs may be important in the pathogenesis of CML. It was
speculated that miR-17, miR-16 and miR-15a/b may be the key
regulators of the MAPK signaling pathway in CML cells.
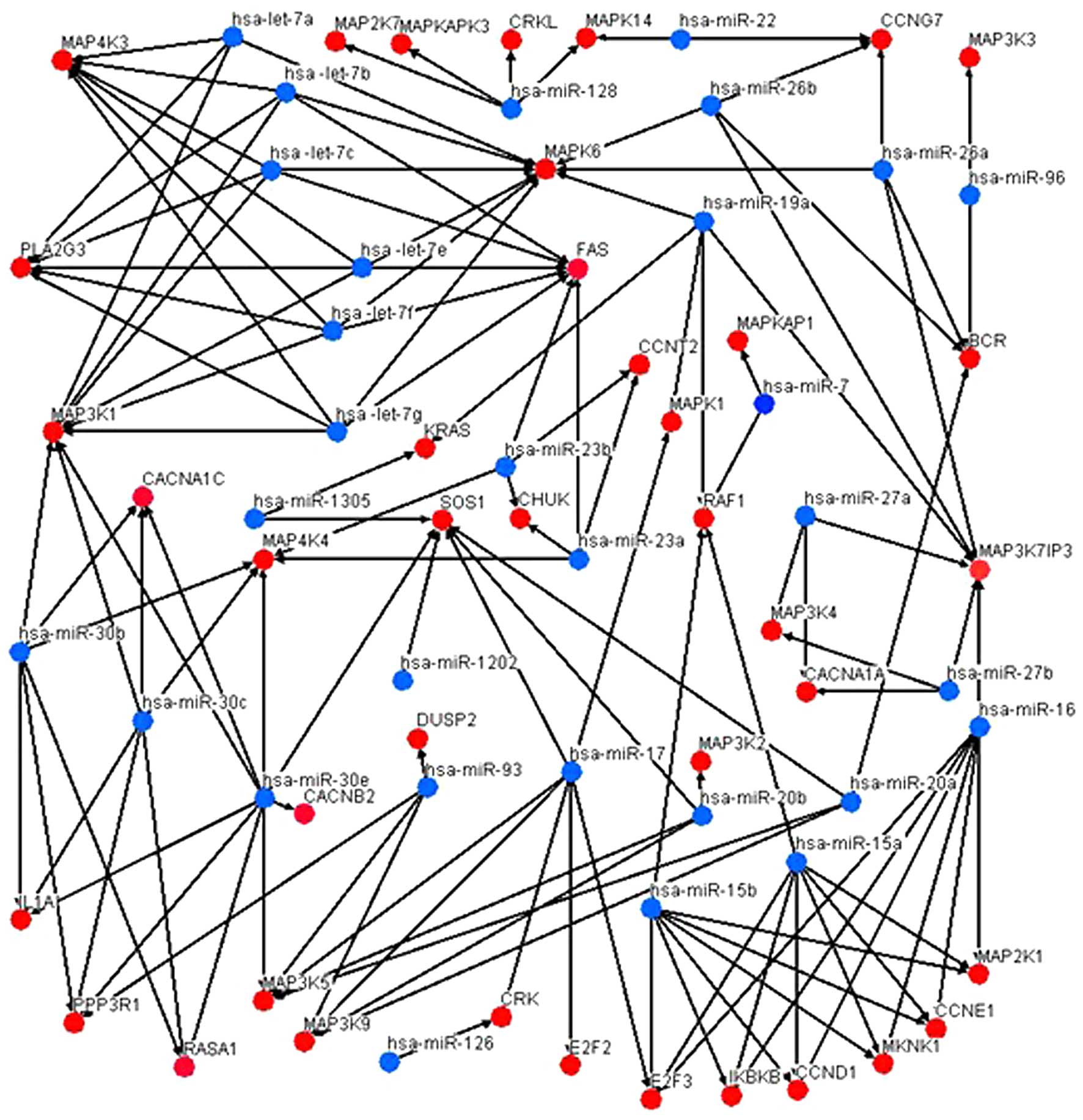 | Figure 4.MicroRNA-gene interaction networks of
the MAPK signaling pathway. Blue nodes represent microRNAs
co-expressed in microvesicles derived from K562 cells and in K562
cells, while red nodes represent their target mRNAs. The edges
represent the regulatory effect of microRNAs on mRNAs. miR-16,
hsa-miR-17 and hsa-miR-15 family (miR-15a and miR-15b) were the
most targeted mRNAs in the MAPK pathway (all exhibited a degree of
7). miRNA, microRNA; mRNA, messenger RNA; hsa, Homo sapiens; MAPK,
mitogen-activated protein kinase; BCR, breakpoint cluster region;
CCNT2, cyclin T2; CACNA1C, calcium voltage-gated channel subunit
alpha1 C; CHUK, conserved helix-loop-helix ubiquitous kinase;
CACNA1A, calcium voltage-gated channel subunit alpha1 A; CACNB2,
calcium voltage-gated channel auxiliary subunit beta 2; CRK, v-crk
avian sarcoma virus CT10 oncogene homolog; CCND1, cyclin D1; CCNE1,
cyclin E1; CCNG7, cyclin G7; DUSP2, dual specificity phosphatase 2;
E2F2, E2F transcription factor 2; E2F3, E2F transcription factor 3;
FAS, Fas cell surface death receptor; IL1A, interleukin 1 alpha;
IKBKB, inhibitor of kappa light polypeptide gene enhancer in
B-cells, kinase beta; KRAS, Kirsten ras oncogene homolog; MAP4K3,
mitogen-activated protein kinase kinase kinase kinase 3; MAP2K7,
mitogen-activated protein kinase kinase 7; MAPKAPK3,
mitogen-activated protein kinase-activated protein kinase 3; CRKL,
CRK like proto-oncogene; MAPK14, mitogen-activated protein kinase
14; MAP3K3, mitogen-activated protein kinase kinase kinase 3;
MAPK6, mitogen-activated protein kinase 6; MAPKAP1,
mitogen-activated protein kinase associated protein 1; MAPK1,
mitogen-activated protein kinase 1; MAP3K1, mitogen-activated
protein kinase kinase kinase 1; MAP4K4, mitogen-activated protein
kinase kinase kinase kinase 4; MAP3K7IP3, mitogen-activated protein
kinase kinase kinase 7-interacting protein 3; MAP3K4,
mitogen-activated protein kinase kinase kinase 4; MAP3K2,
mitogen-activated protein kinase kinase kinase 2; MAP3K5,
mitogen-activated protein kinase kinase kinase 5; MAP3K9,
mitogen-activated protein kinase kinase kinase 9; MKNK1, MAP kinase
interacting serine/threonine kinase 1; MAP2K1, mitogen-activated
protein kinase kinase 1; PLA2G3, phospholipase A2 group III;
PPP3R1, protein phosphatase 3 regulatory subunit B, alpha; RAF1,
v-raf-leukemia viral oncogene 1; RASA1, RAS p21 protein activator
1; SOS1, SOS Ras/Rac guanine nucleotide exchange factor 1 |
Chromosomal localization
A rearrangement (translocation) of genetic material
between chromosomes 9 and 22 is associated with several types of
blood cancer, particularly CML (36,37). As a
typical CML cell line, K562 cells presented t(9:22) (38); thus, the chromosome location of the
differentially expressed microRNAs was further examined. Several
differentially regulated microRNAs in K562-derived MVs, including
miR-27b, miR-24, miR-23b and miR-126*, were located in chromosome
9. A total of 12 co-expressed microRNAs in MVs and their
corresponding parental K562 cells [Homo sapiens (hsa)-let-7a,
hsa-let-7f, miR-126, miR-126*, miR-23b, miR-24, miR-27b and miR-7]
were mapped to chromosome 9, whereas hsa-let-7b, miR-1249, miR-130b
and miR-185 were mapped to chromosome 22. The genomic locations of
a number of these microRNAs were distant from the ABL1 and BCR gene
locus (Fig. 5). Using target
prediction programmes, 875 predicted targets were observed to be
regulated by these 12 microRNAs, which may participate in leukemia
via the above 875 genes. Using TargetScan 5.1 based on the KEGG
pathway database, signaling pathways were analyzed for predicted
targets involved in the MAPK, transforming growth factor (TGF)β,
p53, ErbB, Wnt, vascular endothelial growth factor, Notch and
mechanistic target of rapamycin (mTOR) signaling pathways, focal
adhesion and cell cycle. Table VI
summarizes the predicted targets associated with CML.
 | Table VI.Target genes of microRNAs mapped to
chromosomes 9 and 22 in K562 microvesicles and K562 cells. |
Table VI.
Target genes of microRNAs mapped to
chromosomes 9 and 22 in K562 microvesicles and K562 cells.
| MicroRNAs |
Targetsa | −ln (P-value) | Pathways in
CML |
|---|
| hsa-miR-126 | CRK | 2.13 | 1 |
| miR-24 | CDKN1B, GAB2,
ACVR1B, PIK3R3 | 1.17 | 1, 2, 3 |
| miR-27b | TGFBR1, KRAS, CDK6,
SOS1, RUNX1, GRB2, CBLB, ACVR1C, SHC4, PIK3CD | 4.26 | 1, 2, 4 |
|
hsa-let-7a,hsa-let-7b, | GAB2, TGFBR1,
CDKN1A, CBL, E2F2, RB1, TP53, | 10.85 | 1, 2, 3, 4, 5 |
| hsa-let-7f | AKT2, ACVR1B,
ACVR1C, BCL2L1, CCND |
|
|
| miR-7 | RB1, BCR, PIK3CD,
PIK3R3, BCL2L1, RAF1 | 9.80 | 1, 4, 5 |
| miR-23b | STAT5B, SOS1,
PTPN11, RUNX1, TGFBR2, ACVR1C, PIK3R3, CCND1 | 3.95 | 2 |
| miR-130b | TGFBR1, CDKN1A,
SOS2, E2F2, MAPK1, TGFBR2, ACVR1C, SMAD4 | 4.87 | 1, 2, 4, 6 |
Notably, it was observed that 70 of the 104
microRNAs co-expressed in MVs and in the corresponding parental
K562 cells targeted 184 genes that were mapped to chromosome 9,
while 50 of the 104 microRNAs targeted 66 genes that were mapped to
chromosome 22. These genes were correlated with the cell cycle, p53
signaling pathway, MAPK signaling pathway and RAS oncogene
family.
Discussion
In the present study, K562 MVs microRNA expression
was evaluated, and 7 microRNAs were identified to be significantly
upregulated, while 16 were downregulated, in the K562-MV group
compared with parental K562 cells (Table
I). Those microRNAs may play a role in leukemia. MVs microRNAs
may influence homeostasis (17). MVs
in the leukemia microenvironment may target and fuse with normal
cells, and transfer the dysfunctional microRNA to normal cells,
consequently modifying normal cells in their environment to promote
tumor growth, invasion and metastasis by various mechanisms.
The present study next assessed the microRNA
expression profile of K562 cells. Compared with the microRNA
expression profile of K562-derived MVs and their parental cells,
181/888 microRNAs (20%) were present at elevated levels within MVs
and 226/888 microRNAs (25%) were present at a higher proportion in
the parental cells. In addition, 104 microRNAs were co-expressed
both in MVs and parental cells, which indicated that MVs microRNA
content may, at least in part, reflect the expression of their
parental cells. In other words, the microRNA expression profile of
leukemia-derived MVs is a small version of that exhibited by the
leukemia parental cells.
Notably, 104 co-expressed microRNAs in MVs and their
parental cells included certain microRNA clusters and microRNA
families associated with cancer. For example, the microRNA-23-27-24
cluster is associated with angiogenesis (39), and the oncomir miR-17-92 cluster
(miR-17, miR-19a, miR-19b, miR-20a and miR-92a), which was detected
in CML cluster of differentiation 34+ cells (24), was also observed in the present study.
These clusters encoded seven microRNAs that regulate cell
proliferation, apoptosis and development (40).
Since microRNAs have a global effect on gene
expression, it is not surprising that they may modulate leukemia
progression. In the present study, a number of oncogenes and tumor
suppressor genes were regulated by microRNAs. According to
computational predictions, a single microRNA can target dozens of
genes, and different microRNAs can target one gene (41). The microRNAs displayed co-expression
both in K562 MVs and in the corresponding parental cells,
indicating a common role in leukemia genesis or progression.
It was also observed that 122 elevated microRNAs
were only expressed in parental cells, suggesting that the
compartmentalization of microRNAs from cells into MVs, at least for
certain microRNAs, is an active (selective) process.
The present study also identified target genes
regulated by microRNAs that were dysregulated in MVs and in their
parental K562 cells. It was observed that oncogenes and tumor
suppressor genes were significantly enriched in these target genes.
A large number of the genes identified in the present study as the
potential targets of differentially regulated microRNAs are known
to be involved in cancer through their effects on cell
differentiation [cyclin-dependent kinase 6 and leukemia inhibitory
factor receptor (LIFR)], apoptosis (proviral integration site 1) or
hematopoiesis (GATA binding protein 2 and T-cell acute lymphocytic
leukemia 1). Notably, numerous targets of the cancer-associated
microRNAs, including activin A receptor (ACVR)1B, ACVR1C, Casitas
B-lineage lymphoma (CBL), cyclin (CCN)D1, cyclin-dependent kinase
inhibitor 1B, CT10 regulator of kinase (CRK), cold shock domain
containing E1, ecotropic virus integration site 1 protein homolog,
growth factor receptor-bound protein 2, histone deacetylase 2,
LIFR, phosphoinositide-3-kinase regulatory subunit 3,
phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit
delta, RAF1, retinoblastoma 1, runt related transcription factor 1,
son of sevenless homolog 1 (SOS1), signal transducer and activator
of transcription (STAT)5B, TGFβ receptor (TGFBR)2, TGFBR1 and tumor
protein 53, have been previously reported to be associated with
leukemogenesis (42,43). K562 cells released MVs enriched in
aberrantly expressed microRNAs, including oncomicroRNAs and tumor
suppressor microRNAs, into the leukemia cells microenvironment,
which may play a role in leukemia by abnormally regulating their
target genes, including oncogenes and tumor suppressors,
consequently resulting in leukemia.
The present study also identified signaling pathways
regulated by microRNAs that were dysregulated in MVs and in their
parental K562 cells. A large number of these target genes were
involved in the MAPK, RAS, p53, ErbB, Janus kinase-STAT, TGFβ, mTOR
and Wnt signaling pathways, which are known to be associated with
leukemia (44–50). Consistent with other cancers (51–53), the
role of leukemia-derived MVs was to modulate the disease
progression rather than being the main cause of the disease itself.
MVs from leukemia cells facilitate leukemia progression and
invasion in different ways (15,54–56). MVs
are considered to be taken up by cells through endocytosis and to
release their intravesicular content upon fusing their membrane
with the endosomal membrane (52).
Although the precise mechanisms of uptake are poorly understood, it
is evident that release of microRNAs from the lumen of MVs can
induce activation of specific signal transduction cascades and
influence the physiologic state of recipient cells (52). MVs have an additional advantage over
naturally secreted signaling molecules in that they can present
multiple epitopes to the recipient cell, enabling co-stimulatory
pathways to be activated (57).
Of the predicted targets of microRNAs co-expressed
in MVs and their corresponding parental K562 cells, 30 were
involved in MAPK signaling (including BCR, CBL, CCNE1, CCND1,
CCNT2, CRK, CRK-like, E2F transcription factor (E2F)2, E2F3,
Kirsten RAS, MAPK1, MAPK12, RAF1 and SOS1). The MAPK signaling
pathway is a common point of convergence of various different
mitogenic and anti-apoptotic signal transduction pathways in
hematopoietic and epithelial cancer cells (45). Such deregulation of the MAPK pathway
contributes to BCR-ABL leukemogenesis (58). The MVs derived from leukemia cells may
weaken or enhance the expression of mRNAs involved in the MAPK
pathway in recipient cells, and contribute to the development of
leukemia.
As K562 cells presented t(9:22), several
differentially regulated microRNAs of K562 MVs and their parental
K562 cells were investigated. For example, miR-27b, miR-24, miR-23b
and miR-126* are known to be encoded by the chromosome 9 (59–61). In
the present study, a considerable number of co-expressed microRNAs
in MVs and K562 cells (including hsa-let-7a, hsa-let-7f, miR-126,
miR-126*, miR-23b, miR-24, miR-27b and miR-7) were mapped to
chromosome 9, while let-7b, miR-1249, miR-130b and miR-185 were
mapped to chromosome 22. The genomic locations of various of these
microRNAs were distant from the ABL1 and BCR gene locus (Fig. 5). Alterations in chromosome 9 are
associated with a large number of diseases, particularly cancer
(62). Therefore, there may be a link
between alterations in chromosome 9 and differential expression of
microRNAs.
In summary, the present study first identified the
microRNA expression profile of MVs derived from K562 cells and that
of their parental K562 cells. The fact that 104 microRNAs were
observed to be co-expressed both in MVs and their parental cells
suggests that MVs microRNAs may, at least in part, reflect the
microRNA expression profile of the parental cells. There were
numerous oncogenes, tumour suppressors and signaling pathway genes
that were targeted by these aberrantly expressed MVs microRNAs,
which may contribute to the development of hematopoietic
malignancies. Therefore, leukemia MVs microRNAs may represent a
novel way to intervene therapeutically for treating CML.
Acknowledgements
The present study was supported by the National
Natural Science Foundation of China (Beijing, China; grant no.
81170462). The authors would like to thank the staff at the Center
for Stem Cell Research and Application (Institute of Hematology,
Union Hospital, Tongji Medical College, Huazhong University of
Science and Technology) for their technical assistance and the
staff at the Clinical Laboratory of the Department of Infection
Diseases (Institute of Hematology, Union Hospital, Tongji Medical
College, Huazhong University of Science and Technology) for their
help collecting samples.
References
|
1
|
Zhou JB, Zhang T, Wang BF, Gao HZ and Xu
X: Identification of a novel gene fusion RNF213SLC26A11 in chronic
myeloid leukemia by RNA-Seq. Mol Med Rep. 7:591–597.
2013.PubMed/NCBI
|
|
2
|
Apperley JF: Chronic myeloid leukaemia.
Lancet. 385:1447–1459. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Sasaki K and Mitani K: Molecular
pathogenesis of chronic myeloid leukemia. Nihon Rinsho.
67:1894–1899. 2009.(In Japanese). PubMed/NCBI
|
|
4
|
Belting M and Christianson HC: Role of
exosomes and microvesicles in hypoxia-associated tumour development
and cardiovascular disease. J Intern Med. 278:251–263. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lo Cicero A, Schiera G, Proia P, Saladino
P, Savettieri G, Di Liegro CM and Di Liegro I: Oligodendroglioma
cells shed microvesicles which contain TRAIL as well as molecular
chaperones and induce cell death in astrocytes. Int J Oncol.
39:1353–1357. 2011.PubMed/NCBI
|
|
6
|
Lee TH, D'Asti E, Magnus N, Al-Nedawi K,
Meehan B and Rak J: Microvesicles as mediators of intercellular
communication in cancer-the emerging science of cellular ‘debris’.
Semin Immunopathol. 33:455–467. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Pilzer D, Gasser O, Moskovich O,
Schifferli JA and Fishelson Z: Emission of membrane vesicles: Roles
in complement resistance, immunity and cancer. Springer Semin
Immunopathol. 27:375–387. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
EL Andaloussi S, Mäger I, Breakefield XO
and Wood MJ: Extracellular vesicles: Biology and emerging
therapeutic opportunities. Nat Rev Drug Discov. 12:347–357. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Al-Nedawi K, Meehan B, Micallef J, Lhotak
V, May L, Guha A and Rak J: Intercellular transfer of the oncogenic
receptor EGFRvIII by microvesicles derived from tumour cells. Nat
Cell Biol. 10:619–624. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
10
|
Skog J, Würdinger T, van Rijn S, Meijer
DH, Gainche L, Sena-Esteves M, Curry WT Jr, Carter BS, Krichevsky
AM and Breakefield XO: Glioblastoma microvesicles transport RNA and
proteins that promote tumour growth and provide diagnostic
biomarkers. Nat Cell Biol. 10:1470–1476. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Pisetsky DS, Gauley J and Ullal AJ:
Microparticles as a source of extracellular DNA. Immunol Res.
49:227–234. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Valadi H, Ekström K, Bossios A, Sjöstrand
M, Lee JJ and Lötvall JO: Exosome-mediated transfer of mRNAs and
microRNAs is a novel mechanism of genetic exchange between cells.
Nat Cell Biol. 9:654–659. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
13
|
Taverna S, Flugy A, Saieva L, Kohn EC,
Santoro A, Meraviglia S, De Leo G and Alessandro R: Role of
exosomes released by chronic myelogenous leukemia cells in
angiogenesis. Int J Cancer. 130:2033–2043. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Mineo M, Garfield SH, Taverna S, Flugy A,
De Leo G, Alessandro R and Kohn EC: Exosomes released by K562
chronic myeloid leukemia cells promote angiogenesis in a
Src-dependent fashion. Angiogenesis. 15:33–45. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Szczepanski MJ, Szajnik M, Welsh A,
Whiteside TL and Boyiadzis M: Blast-derived microvesicles in sera
from patients with acute myeloid leukemia suppress natural killer
cell function via membrane-associated transforming growth
factor-beta1. Haematologica. 96:1302–1309. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hunter MP, Ismail N, Zhang X, Aguda BD,
Lee EJ, Yu L, Xiao T, Schafer J, Lee ML, Schmittgen TD, et al:
Detection of microRNA expression in human peripheral blood
microvesicles. PLoS One. 3:e36942008. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Taylor DD and Gercel-Taylor C: MicroRNA
signatures of tumor-derived exosomes as diagnostic biomarkers of
ovarian cancer. Gynecol Oncol. 110:13–21. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wysoczynski M and Ratajczak MZ: Lung
cancer secreted microvesicles: Underappreciated modulators of
microenvironment in expanding tumors. Int J Cancer. 125:1595–1603.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Hayes J, Peruzzi PP and Lawler S:
MicroRNAs in cancer: Biomarkers, functions and therapy. Trends Mol
Med. 20:460–469. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lu J, Getz G, Miska EA, Alvarez-Saavedra
E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA,
et al: MicroRNA expression profiles classify human cancers. Nature.
435:834–838. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Volinia S, Galasso M, Costinean S,
Tagliavini L, Gamberoni G, Drusco A, Marchesini J, Mascellani N,
Sana ME, Abu Jarour R, et al: Reprogramming of miRNA networks in
cancer and leukemia. Genome Res. 20:589–599. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ramkissoon SH, Mainwaring LA, Ogasawara Y,
Keyvanfar K, McCoy JP Jr, Sloand EM, Kajigaya S and Young NS:
Hematopoietic-specific microRNA expression in human cells. Leuk
Res. 30:643–647. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Venturini L, Battmer K, Castoldi M,
Schultheis B, Hochhaus A, Muckenthaler MU, Ganser A, Eder M and
Scherr M: Expression of the miR-17-92 polycistron in chronic
myeloid leukemia (CML) CD34+ cells. Blood. 109:4399–4405. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Taylor DD and Gerçel-Taylor C:
Tumour-derived exosomes and their role in cancer-associated T-cell
signalling defects. Br J Cancer. 92:305–311. 2005.PubMed/NCBI
|
|
25
|
Kosaka N, Iguchi H, Yoshioka Y, Takeshita
F, Matsuki Y and Ochiya T: Secretory mechanisms and intercellular
transfer of MicroRNAs in living Cells. J Biol Chem.
285:17442–17452. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Chen CZ, Li L, Lodish HF and Bartel DP:
MicroRNAs modulate hematopoietic lineage differentiation. Science.
303:83–86. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Li T, Lu YY, Zhao XD, Guo HQ, Liu CH, Li
H, Zhou L, Han YN, Wu KC, Nie YZ, et al: MicroRNA-296-5p increases
proliferation in gastric cancer through repression of
Caudal-related homeobox 1. Onocgene. 33:783–793. 2014. View Article : Google Scholar
|
|
28
|
Li WY, Chen XM, Xiong W, Guo DM, Lu L and
Li HY: Detection of microvesicle miRNA expression in ALL subtypes
and analysis of their functional roles. J Huazhong Univ Sci
Technolog Med Sci. 34:640–645. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Sun L, Hu J, Xiong W, Chen X, Li H and Jie
S: MicroRNA expression profiles of circulating microvesicles in
hepatocellular carcinoma. Acta Gastroenterol Belg. 76:386–392.
2013.PubMed/NCBI
|
|
30
|
Zhang L, Valencia C, Dong B, Chen M, Guan
P and Pan L: Transfer of microRNAs by extracellular membrane
microvesicles: A nascent crosstalk model in tumor pathogenesis,
especially tumor cell-microenvironment interactions. J Hematol
Oncol. 8:142015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Giusti I, D'Ascenzo S and Dolo V:
Microvesicles as potential ovarian cancer biomarkers. Biomed Res
Int. 2013:7030482013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Nieuwland R, Berckmans RJ, McGregor S,
Böing AN, Romijn FP, Westendorp RG, Hack CE and Sturk A: Cellular
origin and procoagulant properties of microparticles in
meningococcal sepsis. Blood. 95:930–935. 2000.PubMed/NCBI
|
|
33
|
Vaz C, Ahmad HM, Sharma P, Gupta R, Kumar
L, Kulshreshtha R and Bhattacharya A: Analysis of microRNA
transcriptome by deep sequencing of small RNA libraries of
peripheral blood. BMC Genomics. 11:2882010. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Linderholm B, Norberg T and Bergh J:
Sequencing of the tumor suppressor gene TP 53. Methods Mol Med.
120:389–401. 2006.PubMed/NCBI
|
|
35
|
Di Bacco A, Keeshan K, McKenna SL and
Cotter TG: Molecular abnormalities in chronic myeloid leukemia:
Deregulation of cell growth and apoptosis. Oncologist. 5:405–415.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Gilbert F: Disease genes and chromosomes:
Disease maps of the human genome. Chromosome 22. Genet Test.
2:89–97. 1998.PubMed/NCBI
|
|
37
|
Gilbert F and Kauff N: Disease genes and
chromosomes: Disease maps of the human genome. Chromosome 9. Genet
Test. 5:157–174. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Chai JH, Zhang Y, Tan WH, Chng WJ, Li B
and Wang X: Regulation of hTERT by BCR-ABL at multiple levels in
K562 cells. BMC Cancer. 11:5122011. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Zhou Q, Gallagher R, Ufret-Vincenty R, Li
X, Olson EN and Wang S: Regulation of angiogenesis and choroidal
neovascularization by members of microRNA-23~27~24 clusters. Proc
Natl Acad Sci USA. 108:8287–8292. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Pospisil V, Vargova K, Kokavec J, Rybarova
J, Savvulidi F, Jonasova A, Necas E, Zavadil J, Laslo P and Stopka
T: Epigenetic silencing of the oncogenic miR-17-92 cluster during
PU.1-directed macrophage differentiation. Embo J. 30:4450–4464.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Lawrie CH, Chi J, Taylor S, Tramonti D,
Ballabio E, Palazzo S, Saunders NJ, Pezzella F, Boultwood J,
Wainscoat JS and Hatton CS: Expression of microRNAs in diffuse
large B cell lymphoma is associated with immunophenotype, survival
and transformation from follicular lymphoma. J Cell Mol Med.
13:1248–1260. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Zhao H, Wang D, Du W, Gu D and Yang R:
MicroRNA and leukemia: Tiny molecule, great function. Crit Rev
Oncol Hematol. 74:149–155. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Cammarata G, Augugliaro L, Salemi D,
Agueli C, La Rosa M, Dagnino L, Civiletto G, Messana F, Marfia A,
Bica MG, et al: Differential expression of specific microRNA and
their targets in acute myeloid leukemia. Am J Hematol. 85:331–339.
2010.PubMed/NCBI
|
|
44
|
Zenz T, Mohr J, Edelmann J, Sarno A, Hoth
P, Heuberger M, Helfrich H, Mertens D, Dohner H and Stilgenbauer S:
Treatment resistance in chronic lymphocytic leukemia: The role of
the p53 pathway. Leuk Lymphoma. 50:510–513. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Wang X, Pesakhov S, Weng A, Kafka M, Gocek
E, Nguyen M, Harrison JS, Danilenko M and Studzinski GP: ERK 5/MAPK
pathway has a major role in 1α,25-(OH)2 vitamin D3-induced terminal
differentiation of myeloid leukemia cells. J Steroid Biochem Mol
Biol. 144:223–227. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Caye A, Strullu M, Guidez F, Cassinat B,
Gazal S, Fenneteau O, Lainey E, Nouri K, Nakhaei-Rad S, Dvorsky R,
et al: Juvenile myelomonocytic leukemia displays mutations in
components of the RAS pathway and the PRC2 network. Nat Genet.
47:1334–1340. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Ufkin ML, Peterson S, Yang X, Driscoll H,
Duarte C and Sathyanarayana P: miR-125a regulates cell cycle,
proliferation, and apoptosis by targeting the ErbB pathway in acute
myeloid leukemia. Leuk Res. 38:402–410. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Hornakova T, Staerk J, Royer Y, Flex E,
Tartaglia M, Constantinescu SN, Knoops L and Renauld JC: Acute
lymphoblastic leukemia-associated JAK1 mutants activate the Janus
kinase/STAT pathway via interleukin-9 receptor alpha homodimers. J
Biol Chem. 284:6773–6781. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Dinner S and Platanias LC: Targeting the
mTOR pathway in Leukemia. J Cell Biochem. 117:1745–1752. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Eaves CJ and Humphries RK: Acute myeloid
leukemia and the Wnt pathway. N Engl J Med. 362:2326–2327. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Malik MF Arshad: Influence of
microvesicles in breast cancer metastasis and their therapeutic
implications. Arch Iran Med. 18:189–192. 2015.PubMed/NCBI
|
|
52
|
Antonyak MA and Cerione RA: Microvesicles
as mediators of intercellular communication in cancer. Methods Mol
Biol. 1165:147–173. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Jorfi S and Inal JM: The role of
microvesicles in cancer progression and drug resistance. Biochem
Soc Trans. 41:293–298. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Wang Y, Cheng Q, Liu J and Dong M:
Leukemia Stem Cell-Released Microvesicles Promote the Survival and
Migration of Myeloid Leukemia Cells and These Effects Can Be
Inhibited by MicroRNA34a Overexpression. Stem Cell Int.
2016:93134252016.
|
|
55
|
Lu L, Chen XM, Tao HM, Xiong W, Jie SH and
Li HY: Regulation of the expression of zinc finger protein genes by
microRNAs enriched within acute lymphoblastic leukemia-derived
microvesicles. Genet Mol Res. 14:11884–11895. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Ghosh AK, Secreto CR, Knox TR, Ding W,
Mukhopadhyay D and Kay NE: Circulating microvesicles in B-cell
chronic lymphocytic leukemia can stimulate marrow stromal cells:
Implications for disease progression. Blood. 115:1755–1764. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
57
|
van der Vos KE, Balaj L, Skog J and
Breakefield XO: Brain tumor microvesicles: insights into
intercellular communication in the nervous system. Cell Mol
Neurobiol. 31:949–959. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Redig AJ, Vakana E and Platanias LC:
Regulation of mammalian target of rapamycin and mitogen activated
protein kinase pathways by BCR-ABL. Leuk Lymphoma. 52:(Suppl 1).
S45–S53. 2011. View Article : Google Scholar
|
|
59
|
Liang T, Yu J, Liu C and Guo L: An
exploration of evolution, maturation, expression and function
relationships in mir-23 ~27 ~24 cluster. PLoS One. 9:e1062232014.
View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Yan J, Ma S, Zhang Y, Yin C, Zhou X and
Zhang G: Potential role of microRNA-126 in the diagnosis of
cancers: A systematic review and meta-analysis. Medicine
(Baltimore). 95:e46442016. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Xiao J, Lin HY, Zhu YY, Zhu YP and Chen
LW: MiR-126 regulates proliferation and invasion in the bladder
cancer BLS cell line by targeting the PIK3R2-mediated PI3K/Akt
signaling pathway. Onco Targets Ther. 9:5181–5193. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Grady B, Goharderakhshan R, Chang J,
Ribeiro-Filho LA, Perinchery G, Franks J, Presti J, Carroll P and
Dahiya R: Frequently deleted loci on chromosome 9 may harbor
several tumor suppressor genes in human renal cell carcinoma. J
Urol. 166:1088–1092. 2001. View Article : Google Scholar : PubMed/NCBI
|