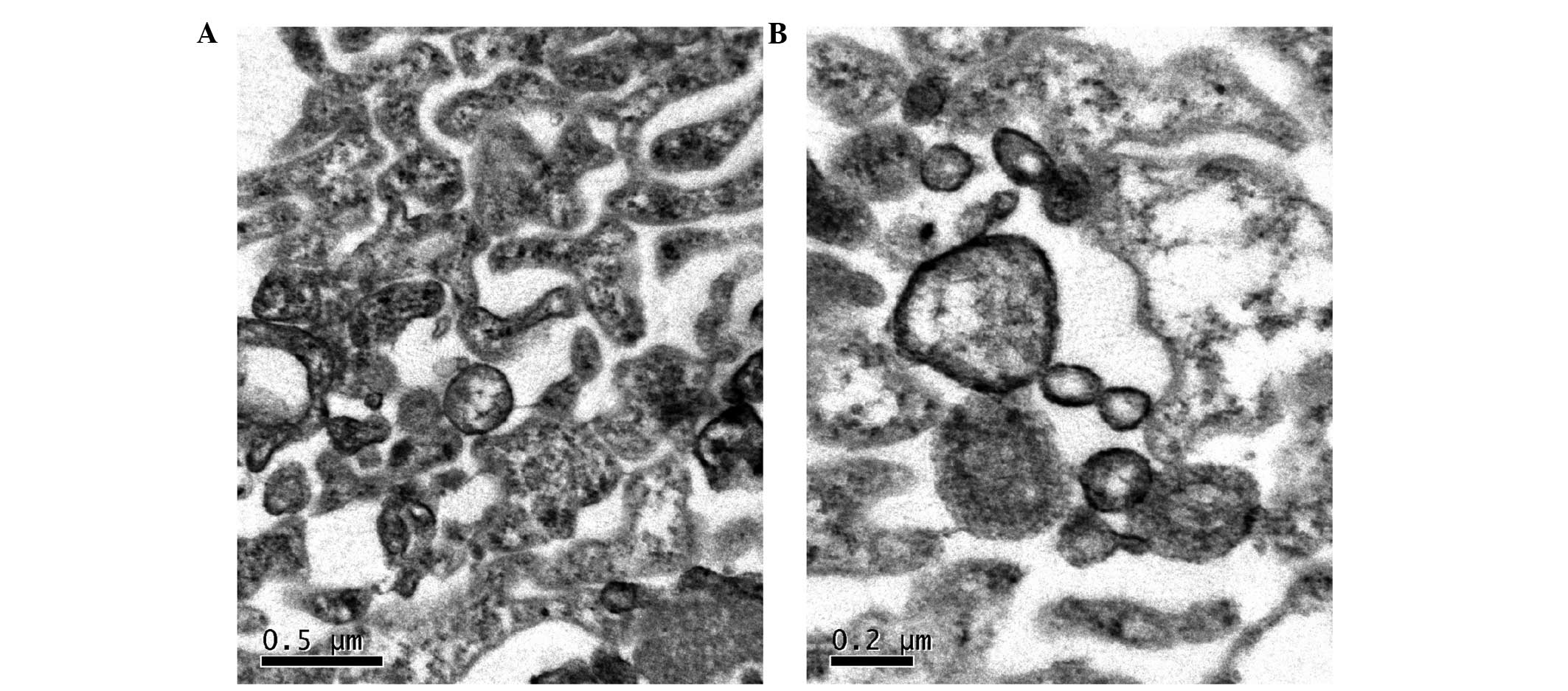
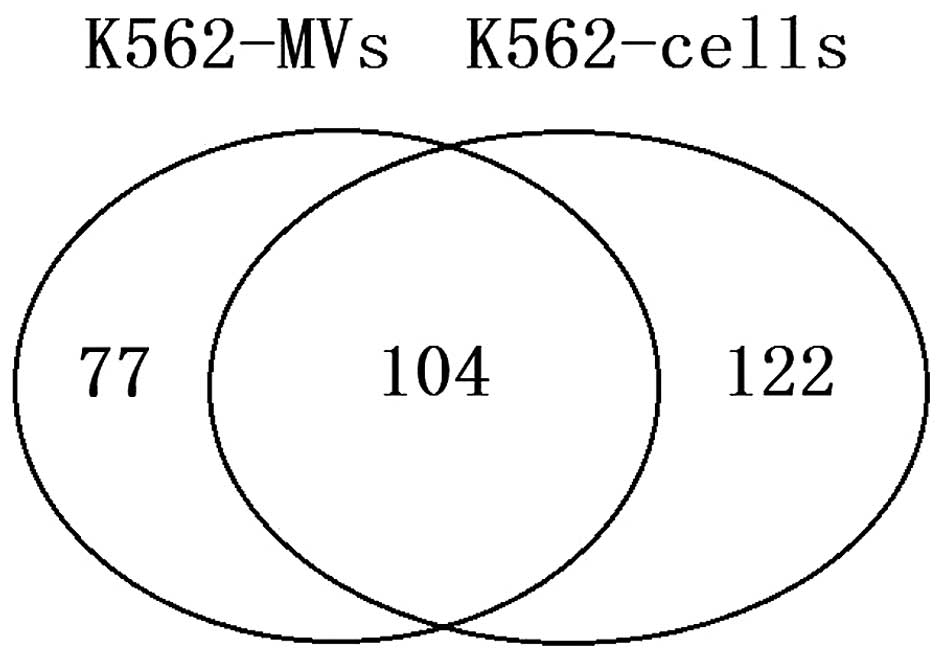
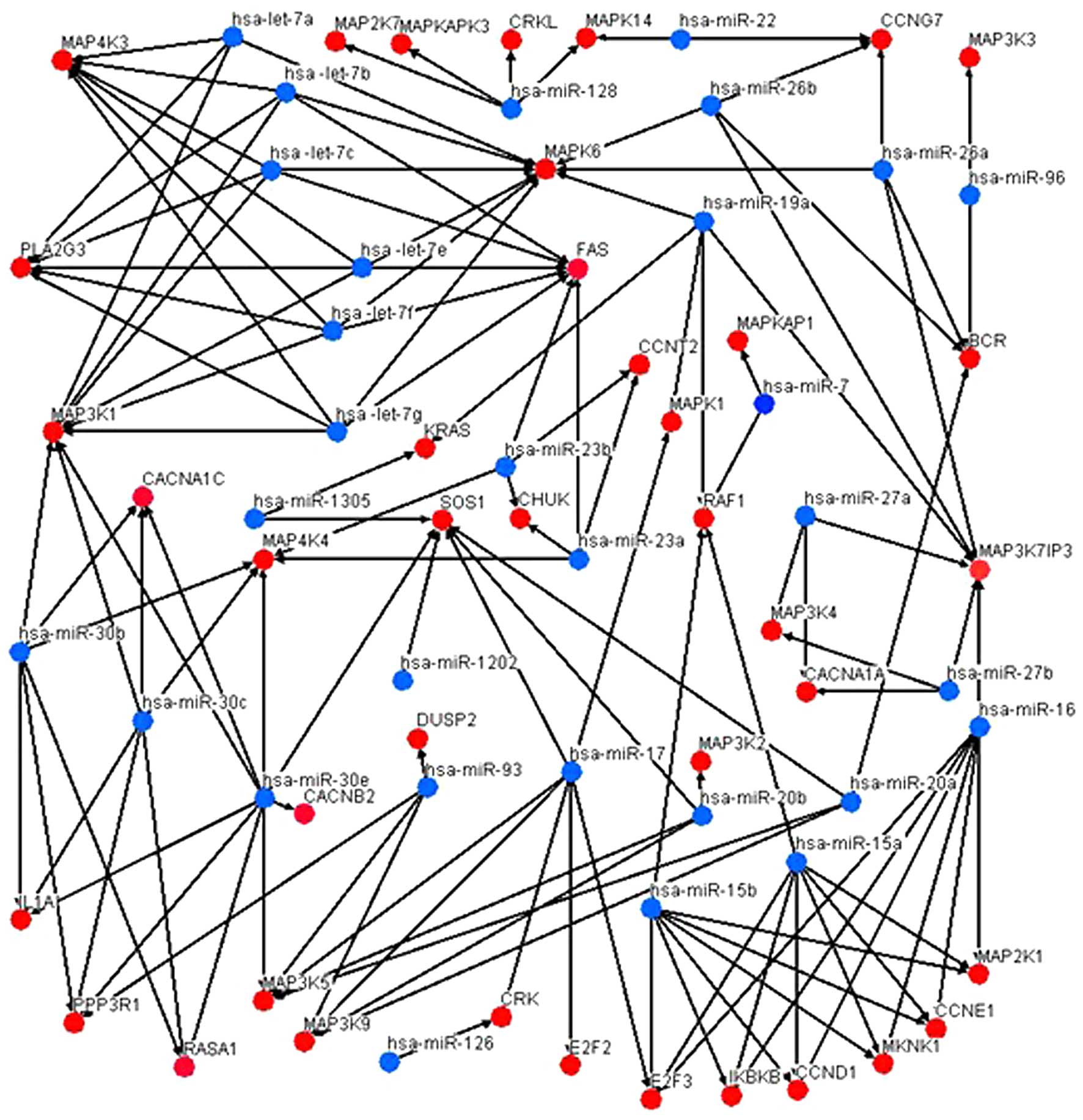
|
1
|
Zhou JB, Zhang T, Wang BF, Gao HZ and Xu
X: Identification of a novel gene fusion RNF213SLC26A11 in chronic
myeloid leukemia by RNA-Seq. Mol Med Rep. 7:591–597.
2013.PubMed/NCBI
|
|
2
|
Apperley JF: Chronic myeloid leukaemia.
Lancet. 385:1447–1459. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Sasaki K and Mitani K: Molecular
pathogenesis of chronic myeloid leukemia. Nihon Rinsho.
67:1894–1899. 2009.(In Japanese). PubMed/NCBI
|
|
4
|
Belting M and Christianson HC: Role of
exosomes and microvesicles in hypoxia-associated tumour development
and cardiovascular disease. J Intern Med. 278:251–263. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lo Cicero A, Schiera G, Proia P, Saladino
P, Savettieri G, Di Liegro CM and Di Liegro I: Oligodendroglioma
cells shed microvesicles which contain TRAIL as well as molecular
chaperones and induce cell death in astrocytes. Int J Oncol.
39:1353–1357. 2011.PubMed/NCBI
|
|
6
|
Lee TH, D'Asti E, Magnus N, Al-Nedawi K,
Meehan B and Rak J: Microvesicles as mediators of intercellular
communication in cancer-the emerging science of cellular ‘debris’.
Semin Immunopathol. 33:455–467. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Pilzer D, Gasser O, Moskovich O,
Schifferli JA and Fishelson Z: Emission of membrane vesicles: Roles
in complement resistance, immunity and cancer. Springer Semin
Immunopathol. 27:375–387. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
EL Andaloussi S, Mäger I, Breakefield XO
and Wood MJ: Extracellular vesicles: Biology and emerging
therapeutic opportunities. Nat Rev Drug Discov. 12:347–357. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Al-Nedawi K, Meehan B, Micallef J, Lhotak
V, May L, Guha A and Rak J: Intercellular transfer of the oncogenic
receptor EGFRvIII by microvesicles derived from tumour cells. Nat
Cell Biol. 10:619–624. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
10
|
Skog J, Würdinger T, van Rijn S, Meijer
DH, Gainche L, Sena-Esteves M, Curry WT Jr, Carter BS, Krichevsky
AM and Breakefield XO: Glioblastoma microvesicles transport RNA and
proteins that promote tumour growth and provide diagnostic
biomarkers. Nat Cell Biol. 10:1470–1476. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Pisetsky DS, Gauley J and Ullal AJ:
Microparticles as a source of extracellular DNA. Immunol Res.
49:227–234. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Valadi H, Ekström K, Bossios A, Sjöstrand
M, Lee JJ and Lötvall JO: Exosome-mediated transfer of mRNAs and
microRNAs is a novel mechanism of genetic exchange between cells.
Nat Cell Biol. 9:654–659. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
13
|
Taverna S, Flugy A, Saieva L, Kohn EC,
Santoro A, Meraviglia S, De Leo G and Alessandro R: Role of
exosomes released by chronic myelogenous leukemia cells in
angiogenesis. Int J Cancer. 130:2033–2043. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Mineo M, Garfield SH, Taverna S, Flugy A,
De Leo G, Alessandro R and Kohn EC: Exosomes released by K562
chronic myeloid leukemia cells promote angiogenesis in a
Src-dependent fashion. Angiogenesis. 15:33–45. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Szczepanski MJ, Szajnik M, Welsh A,
Whiteside TL and Boyiadzis M: Blast-derived microvesicles in sera
from patients with acute myeloid leukemia suppress natural killer
cell function via membrane-associated transforming growth
factor-beta1. Haematologica. 96:1302–1309. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hunter MP, Ismail N, Zhang X, Aguda BD,
Lee EJ, Yu L, Xiao T, Schafer J, Lee ML, Schmittgen TD, et al:
Detection of microRNA expression in human peripheral blood
microvesicles. PLoS One. 3:e36942008. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Taylor DD and Gercel-Taylor C: MicroRNA
signatures of tumor-derived exosomes as diagnostic biomarkers of
ovarian cancer. Gynecol Oncol. 110:13–21. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wysoczynski M and Ratajczak MZ: Lung
cancer secreted microvesicles: Underappreciated modulators of
microenvironment in expanding tumors. Int J Cancer. 125:1595–1603.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Hayes J, Peruzzi PP and Lawler S:
MicroRNAs in cancer: Biomarkers, functions and therapy. Trends Mol
Med. 20:460–469. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lu J, Getz G, Miska EA, Alvarez-Saavedra
E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA,
et al: MicroRNA expression profiles classify human cancers. Nature.
435:834–838. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Volinia S, Galasso M, Costinean S,
Tagliavini L, Gamberoni G, Drusco A, Marchesini J, Mascellani N,
Sana ME, Abu Jarour R, et al: Reprogramming of miRNA networks in
cancer and leukemia. Genome Res. 20:589–599. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ramkissoon SH, Mainwaring LA, Ogasawara Y,
Keyvanfar K, McCoy JP Jr, Sloand EM, Kajigaya S and Young NS:
Hematopoietic-specific microRNA expression in human cells. Leuk
Res. 30:643–647. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Venturini L, Battmer K, Castoldi M,
Schultheis B, Hochhaus A, Muckenthaler MU, Ganser A, Eder M and
Scherr M: Expression of the miR-17-92 polycistron in chronic
myeloid leukemia (CML) CD34+ cells. Blood. 109:4399–4405. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Taylor DD and Gerçel-Taylor C:
Tumour-derived exosomes and their role in cancer-associated T-cell
signalling defects. Br J Cancer. 92:305–311. 2005.PubMed/NCBI
|
|
25
|
Kosaka N, Iguchi H, Yoshioka Y, Takeshita
F, Matsuki Y and Ochiya T: Secretory mechanisms and intercellular
transfer of MicroRNAs in living Cells. J Biol Chem.
285:17442–17452. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Chen CZ, Li L, Lodish HF and Bartel DP:
MicroRNAs modulate hematopoietic lineage differentiation. Science.
303:83–86. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Li T, Lu YY, Zhao XD, Guo HQ, Liu CH, Li
H, Zhou L, Han YN, Wu KC, Nie YZ, et al: MicroRNA-296-5p increases
proliferation in gastric cancer through repression of
Caudal-related homeobox 1. Onocgene. 33:783–793. 2014. View Article : Google Scholar
|
|
28
|
Li WY, Chen XM, Xiong W, Guo DM, Lu L and
Li HY: Detection of microvesicle miRNA expression in ALL subtypes
and analysis of their functional roles. J Huazhong Univ Sci
Technolog Med Sci. 34:640–645. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Sun L, Hu J, Xiong W, Chen X, Li H and Jie
S: MicroRNA expression profiles of circulating microvesicles in
hepatocellular carcinoma. Acta Gastroenterol Belg. 76:386–392.
2013.PubMed/NCBI
|
|
30
|
Zhang L, Valencia C, Dong B, Chen M, Guan
P and Pan L: Transfer of microRNAs by extracellular membrane
microvesicles: A nascent crosstalk model in tumor pathogenesis,
especially tumor cell-microenvironment interactions. J Hematol
Oncol. 8:142015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Giusti I, D'Ascenzo S and Dolo V:
Microvesicles as potential ovarian cancer biomarkers. Biomed Res
Int. 2013:7030482013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Nieuwland R, Berckmans RJ, McGregor S,
Böing AN, Romijn FP, Westendorp RG, Hack CE and Sturk A: Cellular
origin and procoagulant properties of microparticles in
meningococcal sepsis. Blood. 95:930–935. 2000.PubMed/NCBI
|
|
33
|
Vaz C, Ahmad HM, Sharma P, Gupta R, Kumar
L, Kulshreshtha R and Bhattacharya A: Analysis of microRNA
transcriptome by deep sequencing of small RNA libraries of
peripheral blood. BMC Genomics. 11:2882010. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Linderholm B, Norberg T and Bergh J:
Sequencing of the tumor suppressor gene TP 53. Methods Mol Med.
120:389–401. 2006.PubMed/NCBI
|
|
35
|
Di Bacco A, Keeshan K, McKenna SL and
Cotter TG: Molecular abnormalities in chronic myeloid leukemia:
Deregulation of cell growth and apoptosis. Oncologist. 5:405–415.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Gilbert F: Disease genes and chromosomes:
Disease maps of the human genome. Chromosome 22. Genet Test.
2:89–97. 1998.PubMed/NCBI
|
|
37
|
Gilbert F and Kauff N: Disease genes and
chromosomes: Disease maps of the human genome. Chromosome 9. Genet
Test. 5:157–174. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Chai JH, Zhang Y, Tan WH, Chng WJ, Li B
and Wang X: Regulation of hTERT by BCR-ABL at multiple levels in
K562 cells. BMC Cancer. 11:5122011. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Zhou Q, Gallagher R, Ufret-Vincenty R, Li
X, Olson EN and Wang S: Regulation of angiogenesis and choroidal
neovascularization by members of microRNA-23~27~24 clusters. Proc
Natl Acad Sci USA. 108:8287–8292. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Pospisil V, Vargova K, Kokavec J, Rybarova
J, Savvulidi F, Jonasova A, Necas E, Zavadil J, Laslo P and Stopka
T: Epigenetic silencing of the oncogenic miR-17-92 cluster during
PU.1-directed macrophage differentiation. Embo J. 30:4450–4464.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Lawrie CH, Chi J, Taylor S, Tramonti D,
Ballabio E, Palazzo S, Saunders NJ, Pezzella F, Boultwood J,
Wainscoat JS and Hatton CS: Expression of microRNAs in diffuse
large B cell lymphoma is associated with immunophenotype, survival
and transformation from follicular lymphoma. J Cell Mol Med.
13:1248–1260. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Zhao H, Wang D, Du W, Gu D and Yang R:
MicroRNA and leukemia: Tiny molecule, great function. Crit Rev
Oncol Hematol. 74:149–155. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Cammarata G, Augugliaro L, Salemi D,
Agueli C, La Rosa M, Dagnino L, Civiletto G, Messana F, Marfia A,
Bica MG, et al: Differential expression of specific microRNA and
their targets in acute myeloid leukemia. Am J Hematol. 85:331–339.
2010.PubMed/NCBI
|
|
44
|
Zenz T, Mohr J, Edelmann J, Sarno A, Hoth
P, Heuberger M, Helfrich H, Mertens D, Dohner H and Stilgenbauer S:
Treatment resistance in chronic lymphocytic leukemia: The role of
the p53 pathway. Leuk Lymphoma. 50:510–513. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Wang X, Pesakhov S, Weng A, Kafka M, Gocek
E, Nguyen M, Harrison JS, Danilenko M and Studzinski GP: ERK 5/MAPK
pathway has a major role in 1α,25-(OH)2 vitamin D3-induced terminal
differentiation of myeloid leukemia cells. J Steroid Biochem Mol
Biol. 144:223–227. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Caye A, Strullu M, Guidez F, Cassinat B,
Gazal S, Fenneteau O, Lainey E, Nouri K, Nakhaei-Rad S, Dvorsky R,
et al: Juvenile myelomonocytic leukemia displays mutations in
components of the RAS pathway and the PRC2 network. Nat Genet.
47:1334–1340. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Ufkin ML, Peterson S, Yang X, Driscoll H,
Duarte C and Sathyanarayana P: miR-125a regulates cell cycle,
proliferation, and apoptosis by targeting the ErbB pathway in acute
myeloid leukemia. Leuk Res. 38:402–410. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Hornakova T, Staerk J, Royer Y, Flex E,
Tartaglia M, Constantinescu SN, Knoops L and Renauld JC: Acute
lymphoblastic leukemia-associated JAK1 mutants activate the Janus
kinase/STAT pathway via interleukin-9 receptor alpha homodimers. J
Biol Chem. 284:6773–6781. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Dinner S and Platanias LC: Targeting the
mTOR pathway in Leukemia. J Cell Biochem. 117:1745–1752. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Eaves CJ and Humphries RK: Acute myeloid
leukemia and the Wnt pathway. N Engl J Med. 362:2326–2327. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Malik MF Arshad: Influence of
microvesicles in breast cancer metastasis and their therapeutic
implications. Arch Iran Med. 18:189–192. 2015.PubMed/NCBI
|
|
52
|
Antonyak MA and Cerione RA: Microvesicles
as mediators of intercellular communication in cancer. Methods Mol
Biol. 1165:147–173. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Jorfi S and Inal JM: The role of
microvesicles in cancer progression and drug resistance. Biochem
Soc Trans. 41:293–298. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Wang Y, Cheng Q, Liu J and Dong M:
Leukemia Stem Cell-Released Microvesicles Promote the Survival and
Migration of Myeloid Leukemia Cells and These Effects Can Be
Inhibited by MicroRNA34a Overexpression. Stem Cell Int.
2016:93134252016.
|
|
55
|
Lu L, Chen XM, Tao HM, Xiong W, Jie SH and
Li HY: Regulation of the expression of zinc finger protein genes by
microRNAs enriched within acute lymphoblastic leukemia-derived
microvesicles. Genet Mol Res. 14:11884–11895. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Ghosh AK, Secreto CR, Knox TR, Ding W,
Mukhopadhyay D and Kay NE: Circulating microvesicles in B-cell
chronic lymphocytic leukemia can stimulate marrow stromal cells:
Implications for disease progression. Blood. 115:1755–1764. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
57
|
van der Vos KE, Balaj L, Skog J and
Breakefield XO: Brain tumor microvesicles: insights into
intercellular communication in the nervous system. Cell Mol
Neurobiol. 31:949–959. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Redig AJ, Vakana E and Platanias LC:
Regulation of mammalian target of rapamycin and mitogen activated
protein kinase pathways by BCR-ABL. Leuk Lymphoma. 52:(Suppl 1).
S45–S53. 2011. View Article : Google Scholar
|
|
59
|
Liang T, Yu J, Liu C and Guo L: An
exploration of evolution, maturation, expression and function
relationships in mir-23 ~27 ~24 cluster. PLoS One. 9:e1062232014.
View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Yan J, Ma S, Zhang Y, Yin C, Zhou X and
Zhang G: Potential role of microRNA-126 in the diagnosis of
cancers: A systematic review and meta-analysis. Medicine
(Baltimore). 95:e46442016. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Xiao J, Lin HY, Zhu YY, Zhu YP and Chen
LW: MiR-126 regulates proliferation and invasion in the bladder
cancer BLS cell line by targeting the PIK3R2-mediated PI3K/Akt
signaling pathway. Onco Targets Ther. 9:5181–5193. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Grady B, Goharderakhshan R, Chang J,
Ribeiro-Filho LA, Perinchery G, Franks J, Presti J, Carroll P and
Dahiya R: Frequently deleted loci on chromosome 9 may harbor
several tumor suppressor genes in human renal cell carcinoma. J
Urol. 166:1088–1092. 2001. View Article : Google Scholar : PubMed/NCBI
|