Introduction
Colorectal cancer (CRC) is the third most common
cancer type worldwide, with high morbidity and mortality rates
(1). Annually, the global incidence
of CRC is estimated to be ~1 million, with ~500,000 mortalities
(2). Obesity, smoking, diet and a
lack of exercise are risk factors associated with CRC (3). Despite advanced detection approaches,
including colonoscopy and fecal immunochemical testing in early
stage and precancerous lesions (4),
the incidence of CRC remains high. In a previous study, in the
United States in 2014, a cohort of 136,830 individuals was
estimated to be diagnosed with CRC and 50,310 patient (36.8%)
succumbed (5). In China, rapidly
increasing incidence and mortality rates of CRC have been detected
in past decades (6). Therefore,
extensive studies have been conducted to investigate more effective
biological therapies for CRC management. The accumulation of
mutations in a large number of oncogenes and tumor suppressor
genes, which could active or inhibit the pathways critical for the
initiation and progression of CRC, were detected (7). Several biomarkers have been established
for the detection of metastatic CRC, including KRAS and
RAS mutations (8,9). Additionally, the crucial pathways were
also observed. Smith et al showed that tumor protein p53
promoted the progression of CRC through the alteration of genetic
pathways (10). The nuclear factor-κB
signaling pathway was reported to contribute to the carcinogenesis
of CRC (11). MicroRNAs (miRNAs/miRs)
are small RNAs that play central roles in cancer development via
the regulation of its target genes. The altered expression of
miR-21, miR-31, miR-143 and miR-145 was implicated in CRC
progression (12). A recent study
recruiting a genome-wide screening method identified 16 vital genes
in CRC, such as SCARA5, which was affected by methylation
(13). However, the comprehensive
regulatory mechanisms of CRC, particularly the interplayed
associations between miRNAs and genes, remain obscure. The present
study utilized the expression profile data in the study by Khamas
et al (13) to identify the
differentially-expressed genes (DEGs) between CRC tissues and
paired normal control tissues. In addition, the interactions
amongst the DEGs were further investigated through protein-protein
interaction (PPI) network analysis. Furthermore, the miRNAs that
targeted the DEGs were also predicted. As a whole, all these
bioinformatical analyses were aimed to identify potential
biomarkers for the prognosis and prevention of CRC, and to uncover
the underlying regulatory mechanism of CRC progression.
Materials and methods
Gene expression profile data
The gene expression profile data GSE32323, which was
deposited by Khamas et al (13), was used. The public Gene Expression
Omnibus database (http://www.ncbi.nlm.nih.gov/geo/), was utilized in the
study. The platform used was GPL570 (Affymetrix Human Genome U133
Plus 2.0 Array; Agilent Technologies, Palo Alto, CA, USA). In the
expression profile, there were 34 samples derived from the CRC
patients, consisting of 17 from cancerous tissues (CRC samples) and
17 from paired normal tissues (control samples).
Identification of DEGs
Following the data preprocessing, including
background correction and the transformation from probe level to
gene symbol using the Affy package (14) in R language (http://www.bioconductor.org/packages/release/bioc/html/affy.html),
the data was subjected to normalization with the preprocessCore
package (version 1.28.0; http://www.bioconductor.org/packages/3.0/bioc/html/preprocessCore.html)
(15). Subsequently, the DEGs between
CRC and normal samples were selected basing on a t-test of Linear
Models for Microarray Analysis package in R (version 3.22.7;
http://www.bioconductor.org/packages/release/bioc/html/limma.html)
(16). The fold-change (FC) of the
gene expression was also calculated. The threshold criteria for the
DEG selection were P<0.05 and |log2FC| ≥1.
Functional enrichment analysis of the
DEGs
To investigate the functions and processes that may
be altered by the identified DEGs, the Gene ontology (GO) and Kyoto
Encyclopedia of Genes and Genomes pathway enrichment analyses were
performed, using the online tool of the Database for Annotation
Visualization and Integrated Discovery (version 6.7; http://david.abcc.Ncifcrf.gov/) (17), a potent program integrating the gene
or protein functional annotations with graphical summary. The
cut-off value for the screening of significant functions and
pathways was P<0.05.
Establishment of the PPI network
The Search Tool for the Retrieval of Interacting
Genes (STRING) database (version 9.1; http://string-db.org/) (18) was recruited to predict the potential
interactions amongst the identified DEGs from the protein level.
Only the interactions containing at least one DEG were filtered out
to build the PPI network, with the criterion of a combined score of
>0.4, as visualized by Cytoscape (version 3.2.1; http://cytoscape.org/) software (19).
Prediction of targets of
microRNAs
Using the web-based gene set analysis toolkit
(WebGestalt; Vanderbilt University, TN, USA; http://bioinfo.vanderbilt.edu/webgestalt/) (20), the regulatory miRNAs of the DEGs were
selected.
Results
DEGs between CRC and normal
samples
According to the aforementioned selection criteria,
a set of 1,347 DEGs, including 659 upregulated genes and 688
downregulated genes, were identified.
Altered functions and pathways by the
DEGs
As indicated in the results of the enrichment
analysis (Table I), the upregulated
DEGs were significantly enriched in biological processes (BPs) that
included the mitotic cell cycle (GO:0000278), nuclear division
(GO:0000280) and the cell cycle (GO:0007049), and pathways such as
the cell cycle (Hsa04110) and DNA replication (Hsa03030). For the
downregulated DEGs, the over-represented functional GO terms were
cellular response to zinc ion (GO:0071294), cellular response to
chemical stimulus (GO:0070887) and cellular response to chemical
stimulus (GO:0070887), while the prominent pathways were metabolic
pathways (Hsa01100) and pancreatic secretion (Hsa04972) (Table II).
 | Table I.GO and pathway enrichment analysis of
the upregulated DEGs (top 5 in each category, as ranked by the
P-value). |
Table I.
GO and pathway enrichment analysis of
the upregulated DEGs (top 5 in each category, as ranked by the
P-value).
| Category | ID | Term | Count | P-value |
|---|
| BP | GO:0000278 | Mitotic cell
cycle | 102 |
2.63×10−24 |
| BP | GO:0000280 | Nuclear
division | 55 |
2.26×10−22 |
| BP | GO:0007049 | Cell cycle | 128 |
3.47×10−21 |
| BP | GO:0007067 | Mitosis | 55 |
1.25×10−18 |
| BP | GO:0022402 | Cell cycle
process | 117 |
1.15×10−18 |
| CC | GO:0031981 | Nuclear lumen | 140 |
1.68×10−17 |
| CC | GO:0044428 | Nuclear region | 158 |
2.32×10−16 |
| CC | GO:0043233 | Organelle
lumen | 164 |
1.44×10−15 |
| CC | GO:0031974 | Membrane-enclosed
lumen | 166 |
1.55×10−15 |
| CC | GO:0070013 | Intracellular
organelle lumen | 161 |
2.22×10−15 |
| MF | GO:0005515 | Protein
binding | 319 |
2.45×10−8 |
| MF | GO:0005488 | Binding | 450 |
1.92×10−6 |
| MF | GO:0003678 | DNA helicase
activity | 9 |
2.10×10−5 |
| MF | GO:0004386 | Helicase
activity | 15 |
1.42×10−4 |
| MF | GO:0008009 | Chemokine
activity | 8 |
1.70×10−4 |
| KEGG pathway | Hsa04110 | Cell cycle | 24 |
1.21×10−11 |
| KEGG pathway | Hsa03030 | DNA
replication | 11 |
3.64×10−8 |
| KEGG pathway | Hsa03013 | RNA transport | 21 |
1.19×10−7 |
| KEGG pathway | Hsa03008 | Ribosome biogenesis
in eukaryotes | 15 |
1.60×10−7 |
| KEGG pathway | Hsa04115 | p53 signaling
pathway | 10 |
1.72×10−4 |
 | Table II.GO and pathway enrichment analysis of
the downregulated DEGs (top 5 in each category, as ranked by the
P-value). |
Table II.
GO and pathway enrichment analysis of
the downregulated DEGs (top 5 in each category, as ranked by the
P-value).
| Category | ID | Term | Count | P-value |
|---|
| BP | GO:0071294 | Cellular response
to zinc ion | 7 |
2.45×10−8 |
| BP | GO:0070887 | Cellular response
to chemical stimulus | 112 |
2.90×10−7 |
| BP | GO:0010035 | Response to
inorganic substance | 32 |
3.18×10−7 |
| BP | GO:0006629 | Lipid metabolic
process | 77 |
4.91×10−7 |
| BP | GO:0050896 | Response to
stimulus | 303 |
1.34×10−6 |
| CC | GO:0005615 | Extracellular
space | 69 |
6.65×10−11 |
| CC | GO:0005576 | Extracellular
region | 131 |
3.24×10−10 |
| CC | GO:0044421 | Extracellular
region part | 81 |
1.11×10−9 |
| CC | GO:0071944 | Cell periphery | 224 |
1.50×10−9 |
| CC | GO:0016020 | Membrane | 346 |
6.02×10−9 |
| MF | GO:0019955 | Cytokine
binding | 12 |
1.47×10−6 |
| MF | GO:0097367 | Carbohydrate
derivative binding | 20 |
9.75×10−6 |
| MF | GO:0008201 | Heparin
binding | 16 |
1.03×10−5 |
| MF | GO:0005539 | Glycosaminoglycan
binding | 18 |
2.74×10−5 |
| MF | GO:0016616 | Oxidoreductase
activity, acting on the CH-OH group of donors, NAD or NADP as
acceptor | 14 |
3.79×10−5 |
| KEGG pathway | Hsa01100 | Metabolic
pathways | 69 |
1.21×10−4 |
| KEGG pathway | Hsa04972 | Pancreatic
secretion | 12 |
6.96×10−4 |
| KEGG pathway | Hsa04960 |
Aldosterone-regulated sodium
reabsorption | 7 |
1.29×10−3 |
| KEGG pathway | Hsa00910 | Nitrogen
metabolism | 5 |
1.90×10−3 |
| KEGG pathway | Hsa00232 | Caffeine
metabolism | 3 |
2.02×10−3 |
PPI network of the DEGs
By mapping the DEGs into the STRING database, the
potential interactions of the DEGs from the protein level were
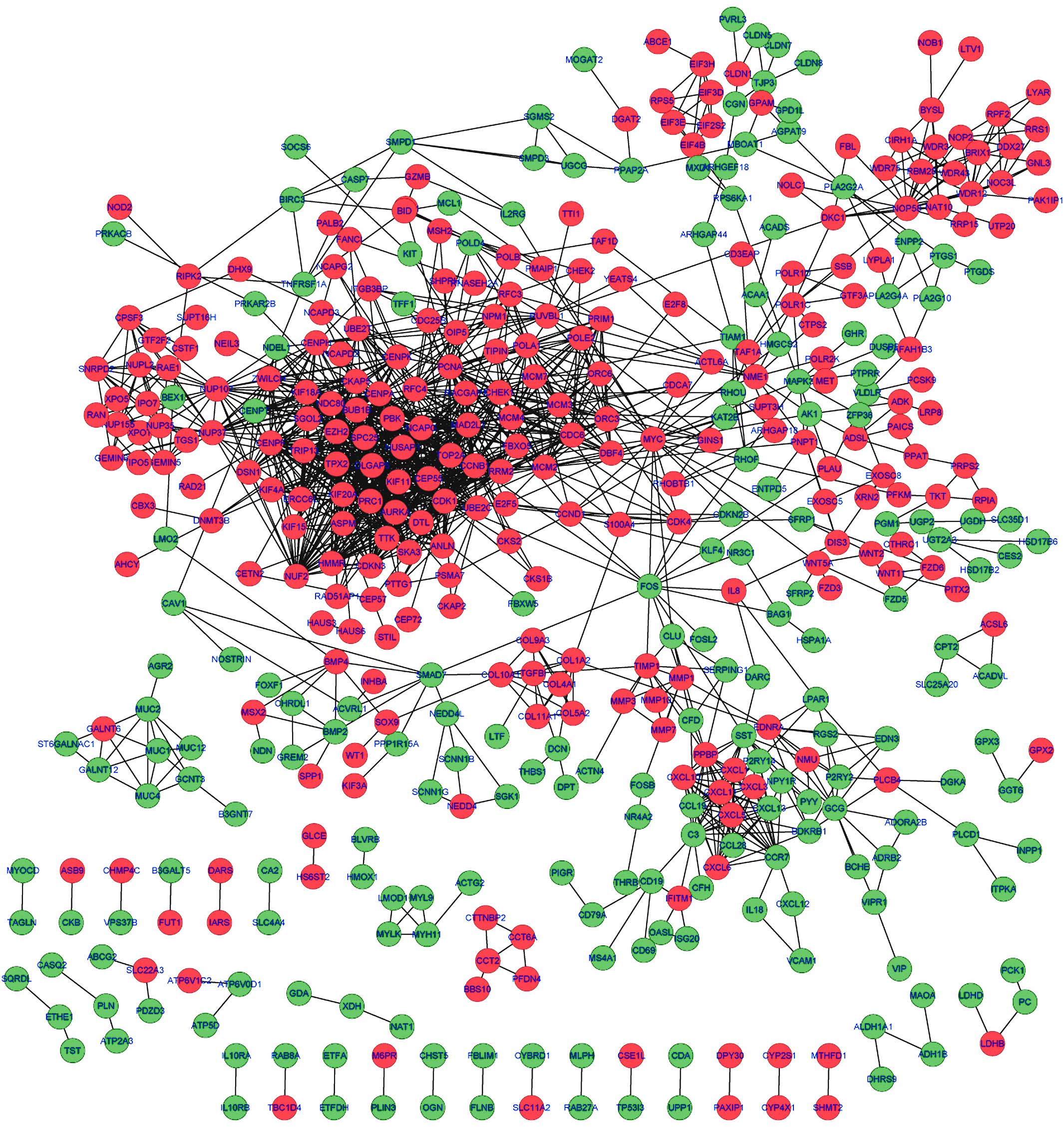
predicted. As a result, a PPI network comprising 1,478 edges and
462 nodes were established. A protein in the network serves as a
‘node’, and the ‘degree’ of a node represents the number of the
interactions between two nodes. Based on this definition, the top
ten nodes with high degrees in the PPI network were
cyclin-dependent kinase 1 (CDK1; degree=59), cyclin B1 (CCNB1;
degree=48), NDC80 kinetochore complex component (degree=45),
non-SMC condensin I complex, subunit G (degree=45), MAD2 mitotic
arrest deficient-like 1 (MAD2L1; degree=44), centromere protein F
(degree=41), BUB1 mitotic checkpoint serine/threonine kinase B
(BUB1B; degree=39), centromere protein A (degree=37), PDZ-binding
kinase (degree=36) and TPX2, microtubule nucleation factor
(degree=36) (Fig. 1).
Integrated miRNA-target regulatory
network
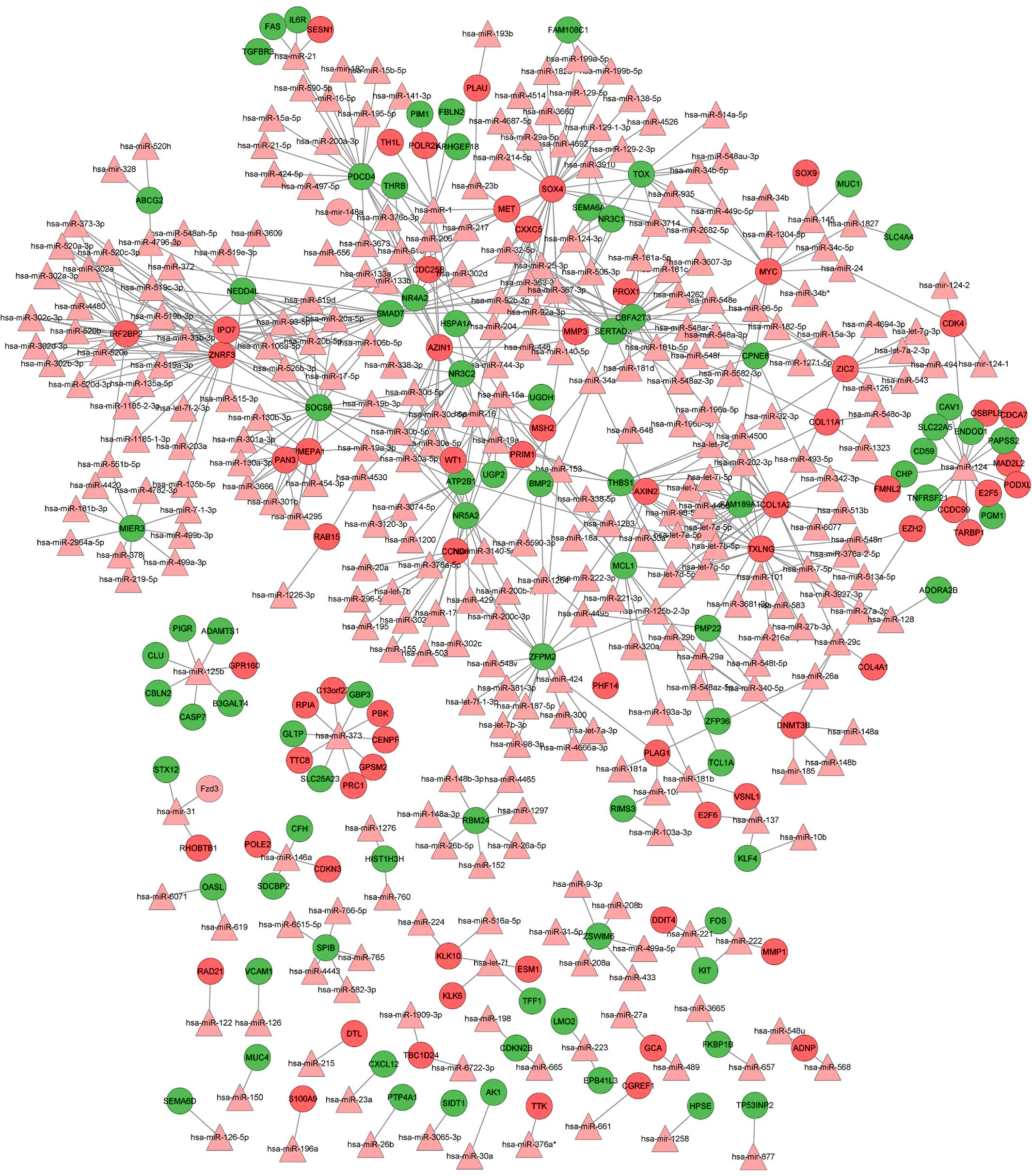
Using the WebGestalt software, the integrated
miRNA-target network was built, consisting of 459 nodes (305 miRNAs
and 154 DEGs) and 646 edges (Fig. 2).
In this network, the notable genes that were targeted by multiple
miRNAs included SRY (sex determining region Y)-box 4 (SOX4;
targeted by 27 miRs, including hsa-mir-129, hsa-mir-133a/b and
hsa-mir-204), CCND1 (cyclin D1; targeted by 21 miRs, including
hsa-let-7b, hsa-mir-155, hsa-mir-16 and hsa-mir-195) and v-myc
avian myelocytomatosis viral oncogene homolog (MYC; targeted by 10
miRs, including hsa-mir-34a, hsa-let-7c, hsa-mir-145 and
hsa-mir-24.
Discussion
CRC is one of the most lethal cancers in the world
(3). Biomarker therapeutic methods
may be the most effective approaches for the management of CRC. In
the present study, a total of 1,347 DEGs (659 upregulated and 688
downregulated) were identified between CRC and normal tissues.
Among them, CDK1, CCNB1, MAD2L1 and
BUB1B, which are mainly enriched in cell cycle-related BPs
and pathways, were also the predominant nodes in the PPI network.
The integrated miRNA-target network identified crucial genes,
including SOX4 (targeted by hsa-mir-129, hsa-mir-133a/b and
hsa-mir-204), MYC (targeted by hsa-mir-34a, hsa-let-7c,
hsa-mir-145 and hsa-mir-24) and CCND1 (targeted by
hsa-let-7b, hsa-mir-155, hsa-mir-16 and hsa-mir-195), which were
all enriched in cell cycle-related pathways. CDK1,
CCNB1 and CCND1 were also associated with the p53
signaling pathways.
Cell cycle-related genes that promote the
proliferation of endothelial cells contribute to the progression of
tumor growth and metastasis of CRC (21). CDK1 encodes for a
serine/threonine kinase that controls the eukaryotic cell cycle by
regulating mitotic onset, as well as the centrosome cycle (22). CDK1 promotes cell proliferation
via phosphorylation and inhibition of forkhead box O1 transcription
factor (23). The alteration of
CDK1 has been found in numerous cancer types, including
breast cancer (24), esophageal
adenocarcinoma (25) and oral
squamous cell carcinoma (26).
Deregulated CDK1 has been found in CRC (27), and it has been demonstrated that
cantharidin, the traditional Chinese medicine that could induce
cell cycle arrest and apoptosis in various cancers, exerted the
anticancer function via the inhibition of CDK1 activity
(28).
CCNB1 is a regulatory protein involved in mitosis
(29). The increased expression of
CCNB1 has also been observed in multiple cancer types,
including non-small cell lung cancer (29) and gastrointestinal stromal tumors
(30). Moreover, CCNB1 serves
as a biomarker for the prognosis of estrogen receptor-positive
breast cancer (31). CCNB1
plays important roles in the cell proliferation at the G2 phase. It
was previously verified that the suppression of CCNB1 by
miR-93 resulted in the inhibition of tumor growth in CRC (32).
MAD2L1 and BUB1B are two major mitotic spindle
checkpoints. Previous studies considered that the mutation or
deficiency in checkpoint proteins may contribute to enhancing the
tumor development in breast cancer (33), and the mutation of BUB1, the paralog
of BUB1B, was first reported in CRC (34). However, in contrast with these
findings, Yuan et al validated the overexpression of
MAD2L1 and BUB1B by reverse
transcription-quantitative polymerase chain reaction in breast
cancer and proposed that it may alternatively be the overexpression
of checkpoint genes that account for genomic instability (35).
The high expression level of SOX4, the
transcription factor responsible for the regulation of embryonic
development and cell control, was significantly associated with the
recurrence of CRC (36). Notably, it
was reported that the oncogene SOX4 was regulated by
miR-129-2 in endometrial cancer, and that the overexpression of
SOX4 was partly caused by the suppression of miR-129-2
(37).
MYC is a central gene that plays important
regulatory roles in cell cycle progression. The deficiency of c-MYC
inhibited the proliferation of tumor cells in numerous cancer types
during the cell cycle through G1 into S phase (38), while the upregulation of MYC
transcription by the SNP rs6983267 was demonstrated to promote the
development of CRC (39). Moreover, a
spectrum of studies has reported the suppression of MYC by miRNAs,
including let-7a (40), miR-23a/b
(41) and miR-145 (42), in various cancer types. Furthermore,
the overexpression of stromal genes, such as collagen type I α2
chain (COL1A2), was also detected in CRC (43).
In the present study, the aforementioned 7 genes
were upregulated in CRC samples, and the genes were all enriched in
cell cycle-related BP terms and pathways, implying that these genes
mediated cell cycle pathways that may play a crucial role in the
tumorigenesis and progression of CRC. Combining the previous
confirmations with the present predicted miRNA-target interactions,
it can be speculated that SOX4 may be the target of miR-129,
while MYC may be targeted by hsa-mir-145 and hsa-let-7c.
The p53 protein acts as a tumor suppressor, as it
could prevent DNA damage by promoting cell cycle arrest in the G1
phase or by apoptosis. The alteration of genes in the p53 signaling
pathway is tightly correlated with cancer development (44) CCND1 is a cyclin protein that functions
as a regulator of CDKs, such as CDK4 or CDK6, during the cell cycle
G1/S transition. Amplification of CCND1 has been observed in
CRC (45) and the association between
increased CCND1 and the activation of the p53 pathway has
been established (46). Besides, the
involvement of CDK1 and CCNB1 in the p53 signaling
pathway have also been implied (47,48). The
present findings indicated that CDK1, CCNB1 and
CCND1 were all enriched in the p53 signaling pathway,
providing a hint that the three genes may have vital roles in the
progression of CRC by the regulation of the p53 signaling pathway.
An extensive number of miRNAs downregulated the expression of
CCND1, including miR-193b (49), miR-200b (50), miR-138b (51) and let-7b (52). Based on the correlations in the
integrated miRNA-target network, CCND1 was regulated by 21
miRNAs, including hsa-let-7c, suggesting that CCND1 may be
the target of hsa-let-7c.
In conclusion, the cell cycle-related pathways
mediated by the CDK1, CCNB1, MAD2L1,
BUB1B, SOX4, COL1A2 and MYC genes, and
the p53 signaling pathway regulated by the CDK1,
CCNB1 and CCND1 genes may play important roles in the
progression of CRC. All these genes may be used as biomarkers for
the prognosis of CRC. Furthermore, SOX4 may be targeted by
miR-129 and MYC by hsa-mir-145 and hsa-let-7c, while
CCND1 may be the target of hsa-let-7c. However, further
experimental validation is warranted to confirm these putative
regulatory correlations.
References
|
1
|
Qiu Y, Patwa TH, Xu L, Shedden K, Misek
DE, Tuck M, Jin G, Ruffin MT, Turgeon DK, Synal S, et al: Plasma
glycoprotein profiling for colorectal cancer biomarker
identification by lectin glycoarray and lectin blot. J Proteome
Res. 7:1693–1703. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Cajuso T, Hänninen UA, Kondelin J, Gylfe
AE, Tanskanen T, Katainen R, Pitkänen E, Ristolainen H, Kaasinen E
and Taipale M: Exome sequencing reveals frequent inactivating
mutations in ARID1A, ARID1B, ARID2 and ARID4A in microsatellite
unstable colorectal cancer. Int J Cancer. 135:611–623. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Mendenhall WM, Amos EH, Rout WR, Zlotecki
RA, Hochwald SN and Cance WG: Adjuvant postoperative radiotherapy
for colon carcinoma. Cancer. 101:1338–1344. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Quintero E, Castells A, Bujanda L,
Cubiella J, Salas D, Lanas Á, Andreu M, Carballo F, Morillas JD,
Hernández C, et al: Colonoscopy versus fecal immunochemical testing
in colorectal-cancer screening. N Engl J Med. 366:697–706. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Siegel R, Desantis C and Jemal A:
Colorectal cancer statistics, 2014. CA Cancer J Clin. 64:104–117.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Huang Z, Huang D, Ni S, Peng Z, Sheng W
and Du X: Plasma microRNAs are promising novel biomarkers for early
detection of colorectal cancer. Int J Cancer. 127:118–126. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Pino MS and Chung DC: The chromosomal
instability pathway in colon cancer. Gastroenterology.
138:2059–2072. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Peeters M, Douillard JY, Van Cutsem E,
Siena S, Zhang K, Williams R and Wiezorek J: Mutant KRAS codon 12
and 13 alleles in patients with metastatic colorectal cancer:
Assessment as prognostic and predictive biomarkers of response to
panitumumab. J Clin Oncol. 31:759–765. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Douillard JY, Oliner KS, Siena S,
Tabernero J, Burkes R, Barugel M, Humblet Y, Bodoky G, Cunningham
D, Jassem J, et al: Panitumumab-FOLFOX4 treatment and RAS mutations
in colorectal cancer. N Engl J Med. 369:1023–1034. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Smith G, Carey FA, Beattie J, Wilkie MJ,
Lightfoot TJ, Coxhead J, Garner RC, Steele RJ and Wolf CR:
Mutations in APC, Kirsten-ras, and p53-alternative genetic pathways
to colorectal cancer. Proc Natl Acad Sci USA. 99:9433–9438. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wang S, Liu Z, Wang L and Zhang X:
NF-kappaB signaling pathway, inflammation and colorectal cancer.
Cell Mol Immunol. 6:327–334. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Slaby O, Svoboda M, Fabian P, Smerdova T,
Knoflickova D, Bednarikova M, Nenutil R and Vyzula R: Altered
expression of miR-21, miR-31, miR-143 and miR-145 is related to
clinicopathologic features of colorectal cancer. Oncology.
72:397–402. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Khamas A, Ishikawa T, Shimokawa K, Mogushi
K, Iida S, Ishiguro M, Mizushima H, Tanaka H, Uetake H and Sugihara
K: Screening for epigenetically masked genes in colorectal cancer
Using 5-Aza-2′-deoxycytidine, microarray and gene expression
profile. Cancer Genomics Proteomics. 9:67–75. 2012.PubMed/NCBI
|
|
14
|
Gautier L, Cope L, Bolstad BM and Irizarry
RA: affy-analysis of Affymetrix GeneChip data at the probe level.
Bioinformatics. 20:307–315. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Bolstad BM: Package ‘preprocessCore’: A
collection of pre-processing functions. R Package version 1.28.0.
http://www.bioconductor.org/packages/3.0/bioc/html/preprocessCore.html2013.Accessed
May 7, 2015.
|
|
16
|
Smyth GK: Limma: Linear models for
microarray dataBioinformatics and Computational Biology Solutions
Using {R} and Bioconductor. Gentleman R, Carey V, Dudoit S,
Irizarry R and Huber W: Springer; New York: pp. 397–420. 2005,
View Article : Google Scholar
|
|
17
|
Dennis G Jr, Sherman BT, Hosack DA, Yang
J, Gao W, Lane HC and Lempicki RA: DAVID: Database for annotation,
visualization, and integrated discovery. Genome Biol. 4:P32003.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Franceschini A, Szklarczyk D, Frankild S,
Kuhn M, Simonovic M, Roth A, Lin J, Minguez P, Bork P, von Mering C
and Jensen LJ: STRING v9.1: Protein-protein interaction networks,
with increased coverage and integration. Nucleic Acids Res.
41:(Database Issue). D808–D815. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wang J, Duncan D, Shi Z and Zhang B:
WEB-based GEne SeT AnaLysis Toolkit (WebGestalt): Update 2013.
Nucleic Acids Res. 41:W77–W83. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Hong BS, Cho JH, Kim H, Choi EJ, Rho S,
Kim J, Kim JH, Choi DS, Kim YK, Hwang D, et al: Colorectal cancer
cell-derived microvesicles are enriched in cell cycle-related mRNAs
that promote proliferation of endothelial cells. BMC Genomics.
10:5562009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Santamaría D, Barrière C, Cerqueira A,
Hunt S, Tardy C, Newton K, Cáceres JF, Dubus P, Malumbres M and
Barbacid M: Cdk1 is sufficient to drive the mammalian cell cycle.
Nature. 448:811–815. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Liu P, Kao TP and Huang H: CDK1 promotes
cell proliferation and survival via phosphorylation and inhibition
of FOXO1 transcription factor. Oncogene. 27:4733–4744. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kim SJ, Nakayama S, Miyoshi Y, Taguchi T,
Tamaki Y, Matsushima T, Torikoshi Y, Tanaka S, Yoshida T, Ishihara
H and Noguchi S: Determination of the specific activity of CDK1 and
CDK2 as a novel prognostic indicator for early breast cancer. Ann
Oncol. 19:68–72. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Hansel DE, Dhara S, Huang RC, Ashfaq R,
Deasel M, Shimada Y, Bernstein HS, Harmon J, Brock M, Forastiere A,
et al: CDC2/CDK1 expression in esophageal adenocarcinoma and
precursor lesions serves as a diagnostic and cancer progression
marker and potential novel drug target. Am J Surg Pathol.
29:390–399. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Chang JT, Wang HM, Chang KW, Chen WH, Wen
MC, Hsu YM, Yung BY, Chen IH, Liao CT, Hsieh LL and Cheng AJ:
Identification of differentially expressed genes in oral squamous
cell carcinoma (OSCC): Overexpression of NPM, CDK1 and NDRG1 and
underexpression of CHES1. Int J Cancer. 114:942–949. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Thoms HC, Dunlop MG and Stark LA:
p38-mediated inactivation of cyclin D1/cyclin-dependent kinase 4
stimulates nucleolar translocation of RelA and apoptosis in
colorectal cancer cells. Cancer Res. 67:1660–1669. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Huang WW, Ko SW, Tsai HY, Chung JG, Chiang
JH, Chen KT, Chen YC, Chen HY, Chen YF and Yang JS: Cantharidin
induces G2/M phase arrest and apoptosis in human colorectal cancer
colo 205 cells through inhibition of CDK1 activity and
caspase-dependent signaling pathways. Int J Oncol. 38:1067–1073.
2011.PubMed/NCBI
|
|
29
|
Soria JC, Jang SJ, Khuri FR, Hassan K, Liu
D, Hong WK and Mao L: Overexpression of cyclin B1 in early-stage
non-small cell lung cancer and its clinical implication. Cancer
Res. 60:4000–4004. 2000.PubMed/NCBI
|
|
30
|
Koon N, Schneider-Stock R, Sarlomo-Rikala
M, Lasota J, Smolkin M, Petroni G, Zaika A, Boltze C, Meyer F,
Andersson L, et al: Molecular targets for tumour progression in
gastrointestinal stromal tumours. Gut. 53:235–240. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Ding K, Li W, Zou Z, Zou X and Wang C:
CCNB1 is a prognostic biomarker for ER+ breast cancer. Med
Hypotheses. 83:359–364. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Yang IP, Tsai HL, Hou MF, Chen KC, Tsai
PC, Huang SW, Chou WW, Wang JY and Juo SH: MicroRNA-93 inhibits
tumor growth and early relapse of human colorectal cancer by
affecting genes involved in the cell cycle. Carcinogenesis.
33:1522–1530. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Scintu M, Vitale R, Prencipe M, Gallo AP,
Bonghi L, Valori VM, Maiello E, Rinaldi M, Signori E, Rabitti C, et
al: Genomic instability and increased expression of BUB1B and
MAD2L1 genes in ductal breast carcinoma. Cancer Lett. 254:298–307.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Shichiri M, Yoshinaga K, Hisatomi H,
Sugihara K and Hirata Y: Genetic and epigenetic inactivation of
mitotic checkpoint genes hBUB1 and hBUBR1 and their relationship to
survival. Cancer Res. 62:13–17. 2002.PubMed/NCBI
|
|
35
|
Yuan B, Xu Y, Woo JH, Wang Y, Bae YK, Yoon
DS, Wersto RP, Tully E, Wilsbach K and Gabrielson E: Increased
expression of mitotic checkpoint genes in breast cancer cells with
chromosomal instability. Clin Cancer Res. 12:405–410. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Andersen CL, Christensen LL, Thorsen K,
Schepeler T, Sørensen FB, Verspaget HW, Simon R, Kruhøffer M,
Aaltonen LA, Laurberg S and Ørntoft TF: Dysregulation of the
transcription factors SOX4, CBFB and SMARCC1 correlates with
outcome of colorectal cancer. Br J Cancer. 100:511–523. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Huang YW, Liu JC, Deatherage DE, Luo J,
Mutch DG, Goodfellow PJ, Miller DS and Huang TH: Epigenetic
repression of microRNA-129-2 leads to overexpression of SOX4
oncogene in endometrial cancer. Cancer Res. 69:9038–9046. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Wang H, Mannava S, Grachtchouk V, Zhuang
D, Soengas MS, Gudkov AV, Prochownik EV and Nikiforov MA: c-Myc
depletion inhibits proliferation of human tumor cells at various
stages of the cell cycle. Oncogene. 27:1905–1915. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Takatsuno Y, Mimori K, Yamamoto K, Sato T,
Niida A, Inoue H, Imoto S, Kawano S, Yamaguchi R, Toh H, et al: The
rs6983267 SNP is associated with MYC transcription efficiency,
which promotes progression and worsens prognosis of colorectal
cancer. Ann Surg Oncol. 20:1395–1402. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Sampson VB, Rong NH, Han J, Yang Q, Aris
V, Soteropoulos P, Petrelli NJ, Dunn SP and Krueger LJ: MicroRNA
let-7a down-regulates MYC and reverts MYC-induced growth in Burkitt
lymphoma cells. Cancer Res. 67:9762–9770. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Gao P, Tchernyshyov I, Chang TC, Lee YS,
Kita K, Ochi T, Zeller KI, De Marzo AM, Van Eyk JE, Mendell JT and
Dang CV: c-Myc suppression of miR-23a/b enhances mitochondrial
glutaminase expression and glutamine metabolism. Nature.
458:762–765. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Chen Z, Zeng H, Guo Y, Liu P, Pan H, Deng
A and Hu J: miRNA-145 inhibits non-small cell lung cancer cell
proliferation by targeting c-Myc. J Exp Clin Cancer Res.
29:1512010. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Smith MJ, Culhane AC, Donovan M, Coffey
JC, Barry BD, Kelly MA, Higgins DG, Wang JH, Kirwan WO, Cotter TG
and Redmond HP: Analysis of differential gene expression in
colorectal cancer and stroma using fluorescence-activated cell
sorting purification. Br J Cancer. 100:1452–1464. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Vazquez A, Bond EE, Levine AJ and Bond GL:
The genetics of the p53 pathway, apoptosis and cancer therapy. Nat
Rev Drug Discov. 7:979–987. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Al-Kuraya K, Novotny H, Bavi P, Siraj AK,
Uddin S, Ezzat A, Sanea NA, Al-Dayel F, Al-Mana H, Sheikh SS, et
al: HER2, TOP2A, CCND1, EGFR and C-MYC oncogene amplification in
colorectal cancer. J Clin Pathol. 60:768–772. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Huang HY, Illei PB, Zhao Z, Mazumdar M,
Huvos AG, Healey JH, Wexler LH, Gorlick R, Meyers P and Ladanyi M:
Ewing sarcomas with p53 mutation or p16/p14ARF homozygous deletion:
A highly lethal subset associated with poor chemoresponse. J Clin
Oncol. 23:548–558. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Choi EJ and Kim GH: Apigenin causes G(2)/M
arrest associated with the modulation of p21(Cip1) and Cdc2 and
activates p53-dependent apoptosis pathway in human breast cancer
SK-BR-3 cells. J Nutr Biochem. 20:285–290. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Li CJ, Li RW, Wang YH and Elsasser TH:
Pathway analysis identifies perturbation of genetic networks
induced by butyrate in a bovine kidney epithelial cell line. Funct
Integr Genomics. 7:193–205. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Chen J, Feilotter HE, Paré GC, Zhang X,
Pemberton JG, Garady C, Lai D, Yang X and Tron VA: MicroRNA-193b
represses cell proliferation and regulates cyclin D1 in melanoma.
Am J Pathol. 176:2520–2529. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Xia W, Li J, Chen L, Huang B, Li S, Yang
G, Ding H, Wang F, Liu N, Zhao Q, et al: MicroRNA-200b regulates
cyclin D1 expression and promotes S-phase entry by targeting RND3
in HeLa cells. Mol Cell Biochem. 344:261–266. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Liu X, Lv XB, Wang XP, Sang Y, Xu S, Hu K,
Wu M, Liang Y, Liu P, Tang J, et al: MiR-138 suppressed
nasopharyngeal carcinoma growth and tumorigenesis by targeting the
CCND1 oncogene. Cell Cycle. 11:2495–2506. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Schultz J, Lorenz P, Gross G, Ibrahim S
and Kunz M: MicroRNA let-7b targets important cell cycle molecules
in malignant melanoma cells and interferes with
anchorage-independent growth. Cell Res. 18:549–557. 2008.
View Article : Google Scholar : PubMed/NCBI
|