Introduction
Breast cancer (BC) is one of the most common cancers
in women worldwide. It is estimated that >508,000 women
succumbed to BC in 2011 (1). Although
BC used to be a common disease in the developed world, recently
~50% of new cases and 58% of deaths have occurred in less developed
countries (1). Localized BC at an
early stage has an improved prognosis and requires less severe
treatment with a survival rate of 98%. However, diagnosis after
tumor metastasis significantly reduces the survival rate to 27%
(2). Early detection may greatly
improve the prognosis of patients with BC. Screening through
mammography has shown a significant reduction of mortality through
the early detection of disease (3).
However, its sensitivity and specificity remain dissatisfactory
(2). False-positive results are more
common for younger women, women who have had previous breast
biopsies, women with a family history of BC and women who are
taking estrogen (4,5). Molecular biomarkers are novel methods of
indirect and direct detection of BC.
Epigenetic alteration is one of the most common
molecular changes identified in the progression of human cancer
(6,7).
Epigenetic mechanisms include aberrant DNA methylation, changes in
histone and chromatin structure by post-translational modification
of histone proteins and alterations in the expression of microRNAs
(8). Aberrant DNA methylation may
alter normal gene expression, genomic structure and genetic
stability (9). It is well established
that widespread changes of DNA methylation occur during
carcinogenesis and tumor progression (10). Distinct from other biomarkers in BC
which are typically based on gene expression, DNA methylation has
been identified to have independent prognostic values that can be
used in tailoring treatment to patients who are receiving uniform
therapy regimens (11).
Promoter methylation predominantly follows a
tumor-specific pattern and has been reported to be a useful
biomarker in various types of cancer, including invasive BC
(12). Previous studies have shown
the frequent methylation of genes involved in cell cycle regulation
[cyclin dependent kinase inhibitor 2A (P16INK4A),
ARF tumor suppressor, cyclin dependent kinase inhibitor 2B, cyclin
D2 (CCDN2) and death associated protein kinase 1
(DAPK)], DNA repair [O-6-methylguanine-DNA methyltransferase
(MGMT) and MutL homolog 1], xenobiotic metabolism
[glutathione S-transferase pi 1 (GSTP1)], signal
transduction [retinoic acid receptor beta 2 (RARβ2),
WNT signaling pathway regulator (APC) and estrogen receptor
2 (ERβ)], adhesion and metastasis (cadherin 1 and cadherin
13) in BC (13–16). As these alterations occurred in cancer
tissues at a higher frequency, they are potentially useful
biomarkers for detecting cancer (17). Using promoter methylation of a panel
of common cancer-related genes can discriminate normal and cancer
tissues with a promising sensitivity and specificity (18). Since BC is heterogeneous, methylation
status and the type of genes remain discordant among different
studies (18). Therefore the true
frequency and utility of DNA methylation as a biomarker in BC has
yet to be established (12).
In order to search for a reliable gene panel for
detecting BC, the present study qualitatively assessed the
methylation frequency of seven candidate genes [breast cancer 1,
early onset; DNA repair associated (BRCA1), GSTP1,
P16INK4A, MGMT, phosphatase and tensin
homolog (PTEN), RARβ2 and CCND2) and
their protein expression in a Chinese population.
Materials and methods
Ethical statement
The Institutional Review Board of Nanjing Medical
University (Nanjing, China) approved the study. Written informed
consent was obtained from all individual participants included in
the study.
Patients and specimens
A total of 70 female patients with BC (age range,
32–93 years; median age, 54.5 years) were recruited from the First
People's Hospital of Zhenjiang City (Zhenjiang, China) between
April 2012 and December 2014. Patients were diagnosed via
pathological evidence. Tumor stage was determined according to the
Classification of Malignant Tumors Staging System (TNM) (19). To investigate the diagnostic value of
DNA methylation in BC, 20 patients with benign breast diseases
(BBD; age range, 20–63 years; median, 40 years) were recruited as
the controls. After obtaining written informed consent from all
participants, trained interviewers conducted a questionnaire to
collect patient's basic characteristics and clinical information.
For patients who had undergone surgery, tissues in the center of
the cancer lesion and remote normal appearing breast tissue were
excised and stored at −80°C until the subsequent extraction of DNA.
None of the enrolled patients had received preoperative
chemotherapy or radiation therapy.
DNA extraction and bisulfite
modification
DNA was extracted from frozen tissue using a QIAamp
DNA Mini kit (Qiagen Inc., Valencia, CA, USA) according to the
manufacturer's instructions. Following quantification via NanoDrop
2000 (Thermo Fisher Scientific, Inc., Waltham, MA, USA), DNA
samples were bisulphite converted and purified using the EpiTect
Fast DNA Bisulfite kit (Qiagen, Inc.).
Primer design and methylation
detection
CpG island methylation at the promoter region of
BRCA1, GSTP1, P16INK4A, MGMT,
PTEN, RARβ2 and CCND2 was determined by
methylation specific polymerase chain reaction (MSP) following
sodium bisulfite modification of the DNA. Prior to the analysis of
the methylation status of the target genes, the presence of
bisulfite modified DNA in each sample was determined by
amplification of 133-bp DNA fragment of the β-actin gene, which was
used for quality control (20).
Modified DNA was amplified in a total volume of 25 µl solution
containing 0.8 U hot-start Taq polymerase (Takara, Japan), 10X PCR
buffer (Mg2+ plus), 2.5 mM of each dNTP, 20 pmol of each
primer and 80 ng of bisulfite-modified genomic DNA as templates.
Cycling conditions consisted of an initial denaturation step at
95°C for 5 min, followed by 38 cycles of 30 sec at 95°C, 30 sec at
the relevant annealing temperature (Table
I) and 45 sec at 72°C. The reaction was terminated with a
10-min extension at 72°C. PCR products (7–8 µl) were resolved on a
2.5% agarose gel containing ethidium bromide and visualized under
UV illumination. To avoid the occurrance of false positive and
false negative results in the reactions, each set of PCR contained
positive and negative controls. Peripheral blood lymphocyte DNA
treated in vitro with SssI methyltransferase (New England
Biolabs, Inc., Beverly, MA, USA) was used as a positive control of
methylated DNA. DNA from normal lymphocytes was used as a control
of unmethylated alleles. PCR reagent without DNA template was used
as a blank control. The primers were designed by Nanjing Steed
BioTechnologies Co., Ltd. (Nanjing, China) (21–27).
 | Table I.Summary of primer sequences,
chromosomal locations, annealing temperatures and product sizes
used for methylation-specific polymerase chain reaction
analyses. |
Table I.
Summary of primer sequences,
chromosomal locations, annealing temperatures and product sizes
used for methylation-specific polymerase chain reaction
analyses.
| Genes | M/U | Sequence
(5′-3′) | Annealing (°C) | Product size
(bp) | Ref |
|---|
| β-actin | – | F:
TGGTGATGGAGGAGGTTTAGTAAGT | 60.0 | 133 | 20 |
|
| – | R:
AACCAATAAAACCTACTCCTCCCTTAA |
|
|
|
| BRCA1 | M | F:
GGTTAATTTAGAGTTTCGAGAGACG | 65.0 | 182 | 21 |
|
|
| R:
TCAACGAACTCACGCCGCGCAATCG |
|
|
|
|
| U | F:
GGTTAATTTAGAGTTTTGAGAGATG | 62.0 | 182 |
|
|
|
| R:
TCAACAAACTCACACCACACAATCA |
|
|
|
| GSTP1 | M | F:
TTCGGGGTGTAGCGGTCGTC | 55.0 | 91 | 22 |
|
|
| R:
GCCCCAATACTAAATCACGACG |
|
|
|
|
| U | F:
GATGTTTGGGGTGTAGTGGTTGTT | 55.0 | 97 |
|
|
|
| R:
CCACCCCAATACTAAATCACAACA |
|
|
|
|
P16INK4A | M | F:
TTATTAGAGGGTGGGGCGGATCGC | 62.0 | 150 | 23 |
|
|
| R:
GACCCCGAACCGCGACCGTAA |
|
|
|
|
| U | F:
TTATTAGAGGGTGGGGTGGATTGT | 60.0 | 151 |
|
|
|
| R:
CAACCCCAAACCACAACCATAA |
|
|
|
| MGMT | M | F:
TTTCGACGTTCGTAGGTTTTCGC | 62.0 | 81 | 24 |
|
|
| R:
GCACTCTTCCGAAAACGAAACG |
|
|
|
|
| U | F:
TTTGTGTTTTGATGTTTGTAGGTTTTTGT | 62.0 | 93 |
|
|
|
| R:
AACTCCACACTCTTCCAAAAACAAAACA |
|
|
|
| PTEN | M | F:
TTCGTTCGTCGTCGTCGTATTT | 62.0 | 207 | 25 |
|
|
| R:
GCCGCTTAACTCTAAACCGCAACCG |
|
|
|
|
| U | F:
GTGTTGGTGGAGGTAGTTGTTT | 62.0 | 163 |
|
|
|
| R:
ACCACTTAACTCTAAACCACAACCA |
|
|
|
| RARβ2 | M | F:
TCGAGAACGCGAGCGATTCG | 62.0 | 146 | 26 |
|
|
| R:
GACCAATCCAACCGAAACGA |
|
|
|
|
| U | F:
TTGAGAATGTGAGTGATTTGA | 60.5 | 146 |
|
|
|
| R:
AACCAATCCAACCAAAACAA |
|
|
|
| CCND2 | M | F:
TCGGTGTGGTTACGTTTAGC | 59.0 | 160 | 27 |
|
|
| R:
TAAAACGACGCGATACAACG |
|
|
|
|
| U | F:
TGGTGTGGTTATGTTTAGTG | 59.0 | 150 |
|
|
|
| R:
ACAATACAACATCTAAAACCAC |
|
|
|
Immunohistochemical analysis
Immunohistochemical analysis was used to evaluate
the expression of specific genes in the breast tissues. Cut from
the paraffin-embedded blocks using a microtome, 4-µm-thick sections
were transferred to gelatin coated slides, and dried at 56°C for 1
h. Paraffin sections on slides were dewaxed in xylene twice for 15
min and rehydrated in a grade series of alcohol (100, 100, 90, 80
and 70%). Slides were subsequently placed in a glass jar filled
with citrate buffer (0.01 M; pH 6.0) in a microwave oven for
antigen retrieval and heated for 10 min at 97°C. Following cooling
in the jar at room temperature, the sections were treated with 3%
H2O2 for 20 min to quench the endogenous
peroxidase activity. Non-specific binding was blocked with 10% goat
serum (ZSJB-BIO, Beijing, China) in phosphate-buffered saline (PBS;
0.01 M; pH 7.4) for 30 min at room temperature. Without rinsing,
the slides were incubated with primary antibodies against
BRCA1 (MS110; ab16780; diluted 1:300 in PBS; Abcam,
Cambridge, UK) and GSTP1 (3F2) (mouse monoclonal; #3369;
diluted 1:800 in PBS; Cell Signaling Technology, Inc., Danvers, MA,
USA) overnight at 4°C. For negative controls, the primary antibody
was replaced by PBS. Slides were washed with PBS, followed by
incubation with the horseradish peroxidase-conjugated secondary
antibody (Polink-2 plus Polymer HRP Detection System; PV-9002;
ZSJB-BIO) for 30 min at room temperature in a moist chamber. The
chromogenic reaction was visualized by diaminobenzidine and
counterstained with hematoxylin. Finally, cells were gradually
dehydrated with graded ethanol and sealed with neutral gum. Images
were captured using an upright fluorescence microscope (Eclipse
80i; Nikon Corporation, Tokyo, Japan).
Immunohistochemical results were scored
independently by two pathologists who were blind to the methylation
status of the samples. The expression of BRCA1 and
GSTP1 was evaluated using the semi-quantitative scoring
criteria according to the staining intensity (0, negative; 1, weak;
2, moderate; and 3, strong) and the proportion of positive cells
(0, positive in ≤5%; 1, positive in >5 and ≤25%; 2, positive in
>25 and ≤50%; 3, positive in >50 and ≤80%; and 4, positive in
>80% tumor cells). The two scores were multiplied together for
each case and gene expression was subsequently graded as: 0,
negative score; 1–4, weak expression score; 5–8, moderate
expression score; and 9–12, strong expression score (28).
Statistical analysis
Pearson's χ2 or Fisher's exact test were
used to compare clinicopathological features between cases and
controls. Mann-Whitney U testing was used for nonparametric
distributed variables. Discriminant validity of selected genes was
examined using the receiver operating characteristic (ROC) curve.
Sensitivity, specificity, and the area under the curve (AUC) were
calculated. P<0.05 was considered to indicate a statistically
significant difference. All statistical analyses were performed
using SPSS 18.0 (SPSS, Inc., Chicago, IL, USA) or STATA 12.0 (Stata
Corporation, College Station, TX, USA).
Results
Promoter methylation in BC and
BBD
A total of 70 patients with BC (age range, 32–93
years; median age, 54.5 years) and 20 patients with BBD (age range,
20–63 years; median, 40 years) were enrolled. The majority of
patients with BC were diagnosed with invasive ductal carcinoma
(82.9%) and 55.7% were defined as stage II. For BBD patients, the
majority were diagnosed with fibroadenoma (75.0%). Promoter
methylation of BRCA1, GSTP1,
P16INK4A, MGMT, PTEN, RARβ2
and CCND2 were measured. The frequency of hypermethylation
in cancer tissues was 24.3, 31.4, 40.0, 27.1, 48.6, 55.7 and 67.1%,
respectively, whereas the frequency of hypermethylation in BBD
tissues was 0.0, 0.0, 20.0, 25.0, 40.0, 40.0 and 45.0%,
respectively. There were 8 (11.4%) cases of hypermethylation in one
gene, 17 (24.3%) cases of hypermethylation in two genes, 14 (20.0%)
cases of hypermethylation in three genes, 17 (24.3%) cases of
hypermethylation in four genes, 6 (8.6%) cases of hypermethylation
in five genes, and 4 (5.7%) cases of hypermethylation in six genes.
Only four patients did not exhibit any hypermethylation in these
seven genes. BRCA1 (24.3% in BC vs. 0.0% in BBD; P=0.034)
and GSTP1 (31.4% in BC vs. 0.0% in BBD; P=0.010) were
significantly hypermethylated in BC as compared with BBD controls
(Table II). Fig. 1 summarizes the methylation patterns of
selected genes.
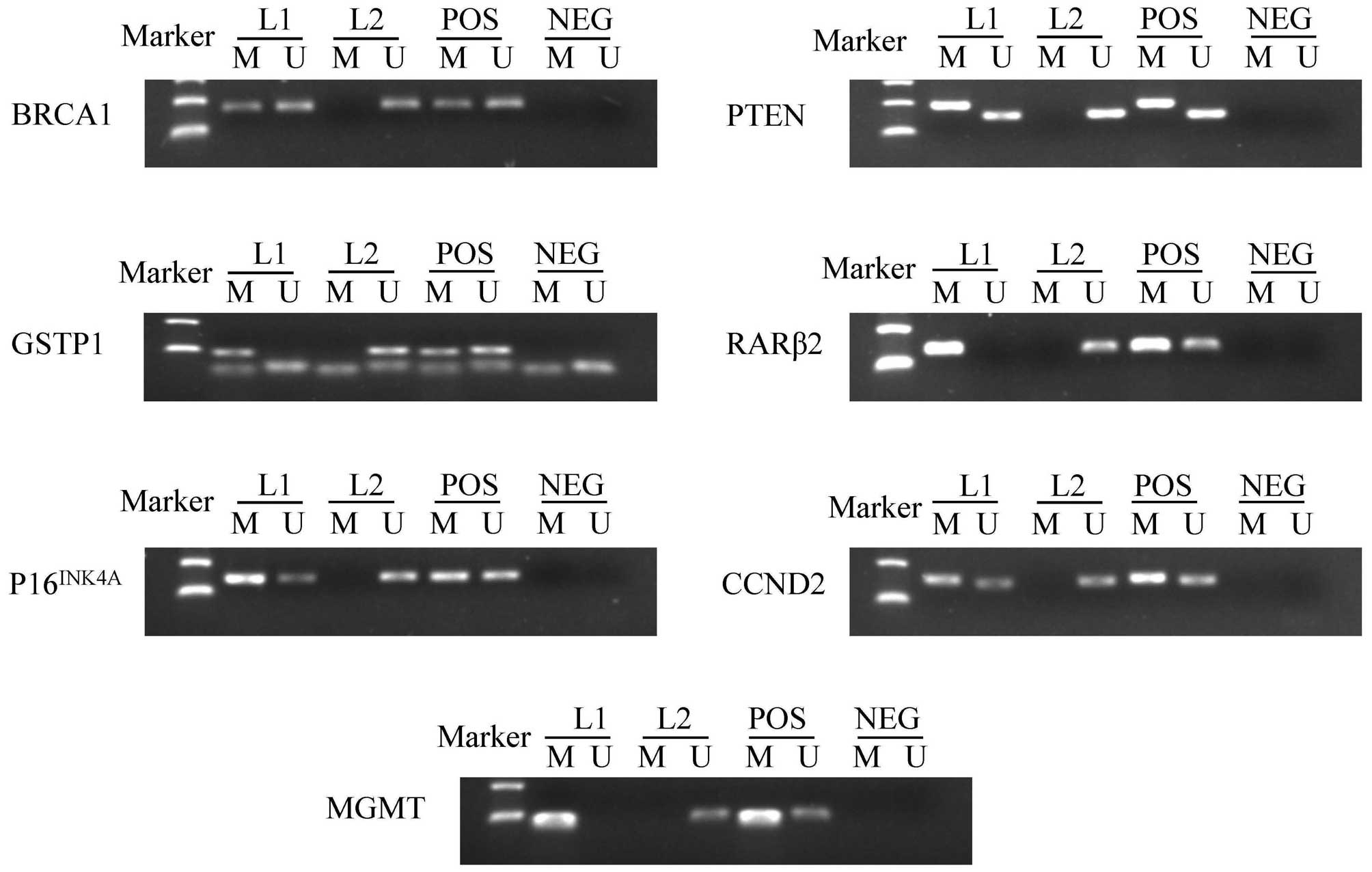 | Figure 1.Representative results of
methylation-specific polymerase chain reaction analyses. Peripheral
blood lymphocytes DNA treated by SssI methyltransferase was used as
the methylation-positive control and DNA from normal lymphocytes
was used as the unmethylation-positive control. NEG, negative
control; Lane L1-L2, breast cancer samples; M, methylation; U,
unmethylation; POS, positive control; BRCA1, breast cancer
1, early onset; DNA repair associated; GSTP1, glutathione
S-transferase pi 1; P16INK4A, cyclin dependent
kinase inhibitor 2A; MGMT, O-6-methylguanine-DNA
methyltransferase; PTEN, phosphatase and tensin homolog;
RARβ2, retinoic acid receptor beta 2; CCND2, cyclin
D2. |
 | Table II.Methylation status of patients with
BC and BBD. |
Table II.
Methylation status of patients with
BC and BBD.
| Genes | MU | BC, n (%) | BBD, n (%) | P-value |
|---|
| BRCA1 | M | 17 (24.3) | 0 (0.0) | 0.034 |
|
| U | 53 (75.7) | 20 (100.0) |
|
| GSTP1 | M | 22 (31.4) | 0 (0.0) | 0.010 |
|
| U | 48 (68.6) | 20 (100.0) |
|
|
P16INK4A | M | 28 (40.0) | 4 (20.0) | 0.099 |
|
| U | 42 (60.0) | 16 (80.0) |
|
| MGMT | M | 19 (27.1) | 5 (25.0) | 0.848 |
|
| U | 51 (72.9) | 15 (75.0) |
|
| PTEN | M | 34 (48.6) | 8 (40.0) | 0.498 |
|
| U | 36 (51.4) | 12 (60.0) |
|
| RARβ2 | M | 39 (55.7) | 8 (40.0) | 0.215 |
|
| U | 31 (44.3) | 12 (60.0) |
|
| CCND2 | M | 47 (67.1) | 9 (45.0) | 0.072 |
|
| U | 23 (32.9) | 11 (55.0) |
|
The sensitivity and specificity of each gene in
distinguishing BC was calculated (Table
III). The AUC for selected genes ranged from 0.511 to 0.657.
The sensitivity of each gene ranged from 24.3 to 67.1% and the
specificity ranged from 55.0 to 100.0%. Methylation was scored as 1
and unmethylation as 0. The scores of the selected genes of the
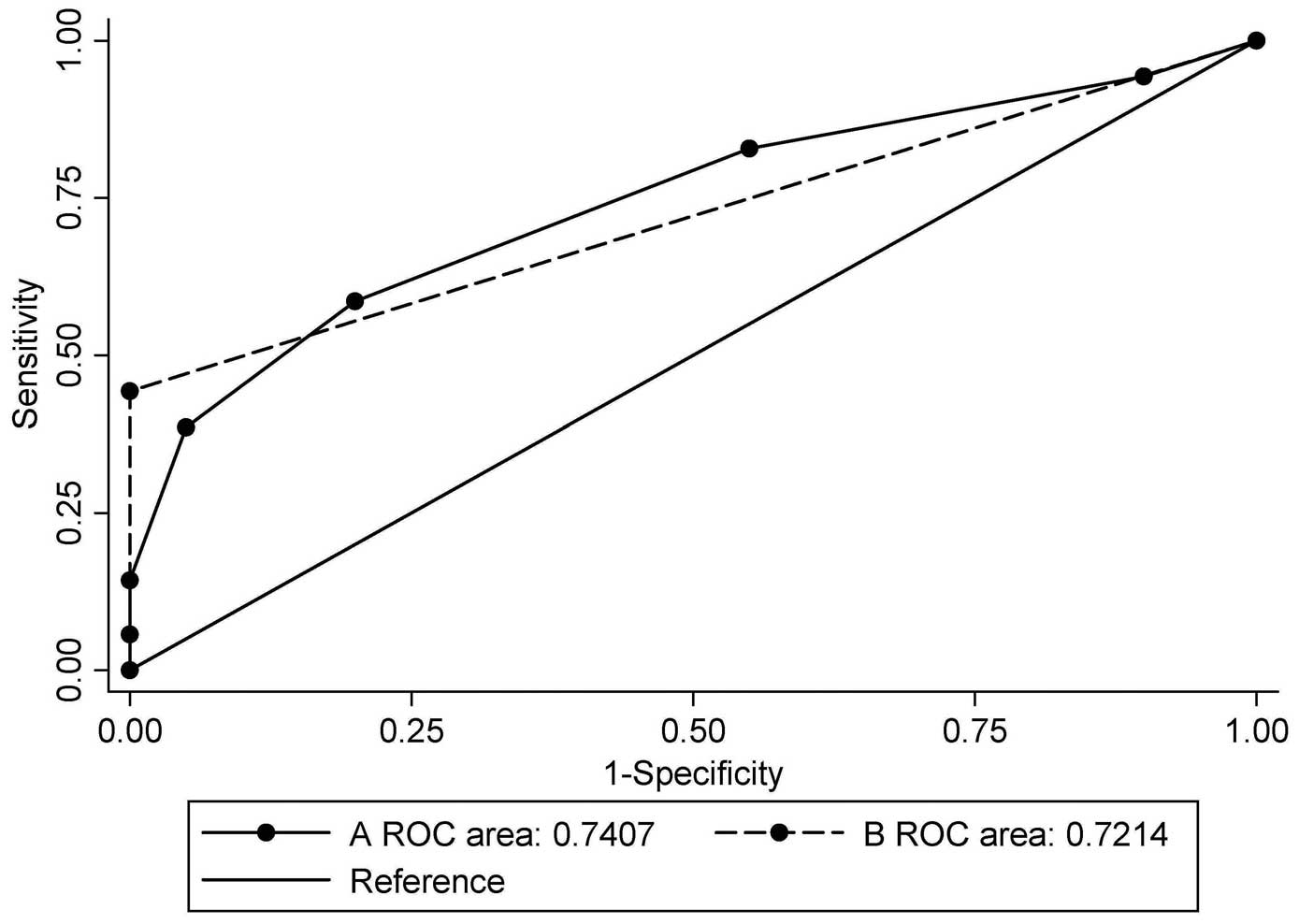
biomarker were totalled. When the combination of BRCA1 and
GSTP1 was used, the AUC was 0.721 [95% confidence interval
(CI), 0.616–0.827; P=0.003], with a sensitivity of 44.3% and a
specificity of 100.0% at the cut-off point of 1, which indicated
hypermethylation in at least one gene (Table IV). When all seven candidate genes
were used, the AUC was 0.741 (95% CI, 0.631–0.850; P=0.001), with a
sensitivity of 58.6% and a specificity of 80.0% when the cut-off
point was set at 3, which indicated hypermethylation in at least
three genes (Table V). Fig. 2 illustrates the ROC curves of
different combinations.
 | Table III.Diagnostic performance of candidate
genes. |
Table III.
Diagnostic performance of candidate
genes.
| Gene | BC pos./total | BBD pos./total | Sensitivity
(%) | Specificity
(%) | AUC | 95% CI | P-value |
|---|
| BRCA1 | 17/70 | 0/20 | 24.3 | 100.0 | 0.621 | 0.497–0.745 | 0.099 |
| GSTP1 | 22/70 | 0/20 | 31.4 | 100.0 | 0.657 | 0.540–0.775 | 0.033 |
|
P16INK4A | 28/70 | 4/20 | 40.0 |
80.0 | 0.600 | 0.465–0.735 | 0.174 |
| MGMT | 19/70 | 5/20 | 27.1 |
75.0 | 0.511 | 0.367–0.654 | 0.884 |
| PTEN | 34/70 | 8/20 | 48.6 |
60.0 | 0.543 | 0.400–0.686 | 0.560 |
| RARβ2 | 39/70 | 8/20 | 55.7 |
60.0 | 0.579 | 0.437–0.720 | 0.286 |
| CCND2 | 47/70 | 9/20 | 67.1 |
55.0 | 0.611 | 0.468–0.754 | 0.133 |
 | Table IV.Combination of BRCA1 and
GSTP1 for the diagnosis of breast cancer. |
Table IV.
Combination of BRCA1 and
GSTP1 for the diagnosis of breast cancer.
| Cut-point | Sensitivity
(%) | Specificity
(%) | Correctly
classified (%) | LR+ | LR- |
|---|
| ≥0 | 100.0 |
0.0 | 77.8 | 1.00 | – |
| ≥1 |
44.3 | 100.0 | 56.7 | – | 0.56 |
| ≥2 |
11.4 | 100.0 | 31.1 | – | 0.89 |
| >2 |
0.0 | 100.0 | 22.2 | – | 1.00 |
 | Table V.Combination of seven candidate genes
for the diagnosis of breast cancer. |
Table V.
Combination of seven candidate genes
for the diagnosis of breast cancer.
| Cut-point | Sensitivity
(%) | Specificity
(%) | Correctly
classified (%) | LR+ | LR- |
|---|
| ≥0 | 100.0 |
0.0 | 77.8 | 1.00 | – |
| ≥1 |
94.3 |
10.0 | 75.6 | 1.05 | 0.57 |
| ≥2 |
82.9 |
45.0 | 74.4 | 1.51 | 0.38 |
| ≥3 |
58.6 |
80.0 | 63.3 | 2.93 | 0.52 |
| ≥4 |
38.6 |
95.0 | 51.1 | 7.71 | 0.65 |
| ≥5 |
14.3 | 100.0 | 33.3 | – | 0.86 |
| ≥6 |
5.7 | 100.0 | 26.7 | – | 0.94 |
| >6 |
0.0 | 100.0 | 22.2 | – | 1.00 |
Association of methylation status and
clinicopathological parameters
Table VI presents the
methylation status of patients included in the present study
stratified by age, tumor size, histologic type, clinical stage,
lymph node metastases, menopausal status and the expression levels
of estrogen receptor (ER), progesterone receptor
(PR), human epidermal growth factor receptor 2 (HER2)
and P53 in cancerous tissues. Hypermethylation of
BRCA1 was demonstrated to be significantly more frequent in
patients with lymph node metastasis (P=0.025). Hypermethylation of
P16INK4A was significantly associated with age
(P=0.015), menopausal status (P=0.003) and P53 expression
(P=0.011). Hypermethylation of PTEN was significantly
associated with menopausal status (P=0.027). RARβ2
hypermethylation was significantly more common in
ER-negative (P=0.002), PR-negative (P<0.001) and
P53-positive tumors (P=0.020).
 | Table VI.Correlation of DNA methylation and
clinicopathological parameters. |
Table VI.
Correlation of DNA methylation and
clinicopathological parameters.
|
|
| BRCA1
(%) | GSTP1
(%) |
P16INK4A (%) | MGMT
(%) | PTEN
(%) | RARβ2
(%) | CCND2
(%) |
|---|
|
|
|
|
|
|
|
|
|
|
|---|
|
Characteristics | N (%) | M | U | M | U | M | U | M | U | M | U | M | U | M | U |
|---|
| Age (years) |
|
|
<55 | 35 (50.0) | 10 (28.6) | 25 (71.4) | 8 (22.9) | 27 (77.1) | 9 (25.7) | 26 (74.3) | 9 (25.7) | 26 (74.3) | 14 (40.0) | 21 (60.0) | 19 (54.3) | 16 (45.7) | 22 (62.9) | 13 (37.1) |
|
≥55 | 35 (50.0) | 7 (20.0) | 28 (80.0) | 14 (40.0) | 21 (60.0) | 19 (54.3) | 16 (45.7) | 10 (28.6) | 25 (71.4) | 20 (57.1) | 15 (42.9) | 20 (57.1) | 15 (42.9) | 25 (71.4) | 10 (28.6) |
|
P-value |
| 0.403 |
| 0.122 |
| 0.015 |
| 0.788 |
| 0.151 |
| 0.810 |
| 0.445 |
|
| Size of tumor |
|
| <2
cm | 18 (25.7) | 5 (27.8) | 13 (72.2) | 6 (33.3) | 12 (66.7) | 8 (44.4) | 10 (55.6) | 6 (33.3) | 12 (66.7) | 9 (50.0) | 9 (50.0) | 9 (50.0) | 9 (50.0) | 13 (72.2) | 5 (27.8) |
| ≥2
cm | 52 (74.3) | 12 (23.1) | 40 (76.9) | 16 (30.8) | 36 (69.2) | 20 (38.5) | 32 (61.5) | 13 (25.0) | 39 (75.0) | 25 (48.1) | 27 (51.9) | 30 (57.7) | 22 (42.3) | 34 (65.4) | 18 (34.6) |
|
P-value |
| 0.935 |
| 0.840 |
| 0.655 |
| 0.706 |
| 0.888 |
| 0.571 |
| 0.594 |
|
| Histologic
type |
|
|
DCIS | 5 (7.1) | 2 (40.0) | 3 (60.0) | 2 (40.0) | 3 (60.0) | 4 (80.0) | 1 (20.0) | 2 (40.0) | 3 (60.0) | 4 (80.0) | 1 (20.0) | 3 (60.0) | 2 (40.0) | 3 (60.0) | 2 (40.0) |
|
IDC | 58 (82.9) | 15 (25.9) | 43 (74.1) | 19 (32.8) | 39 (67.2) | 21 (36.2) | 37 (63.8) | 17 (29.3) | 41 (70.7) | 26 (44.8) | 32 (55.2) | 33 (56.9) | 25 (43.1) | 39 (67.2) | 19 (32.8) |
|
Others | 7 (10.0) | 0 (0.0) | 7 (100.0) | 1 (14.3) | 6 (85.7) | 3 (42.9) | 4 (57.1) | 0 (0.0) | 7 (100.0) | 4 (57.1) | 3 (42.9) | 3 (42.9) | 4 (57.1) | 5 (71.4) | 2 (28.6) |
|
P-value |
| 0.202 |
| 0.602 |
| 0.151 |
| 0.173 |
| 0.289 |
| 0.891 |
| 1.00 |
|
| Clinical
stages |
|
| I | 13 (18.6) | 3 (23.1) | 10 (76.9) | 4 (30.8) | 9 (69.2) | 5 (38.5) | 8 (61.5) | 4 (30.8) | 9 (69.2) | 7 (53.8) | 6 (46.2) | 6 (46.2) | 7 (53.8) | 9 (69.2) | 4 (30.8) |
| II | 39 (55.7) | 10 (25.6) | 29 (74.4) | 11 (28.2) | 28 (71.8) | 13 (33.3) | 26 (66.7) | 8 (20.5) | 31 (79.5) | 18 (46.2) | 21 (53.8) | 23 (59.0) | 16 (41.0) | 25 (64.1) | 14 (35.9) |
|
III | 18 (25.7) | 4 (22.2) | 14 (77.8) | 7 (38.9) | 11 (61.1) | 10 (55.6) | 8 (44.4) | 7 (38.9) | 11 (61.1) | 9 (50.0) | 9 (50.0) | 10 (55.6) | 8 (44.4) | 13 (72.2) | 5 (27.8) |
|
P-value |
| 0.955 |
| 0.725 |
| 0.279 |
| 0.338 |
| 0.882 |
| 0.723 |
| 0.817 |
|
| Lymph node
metastases |
|
| No | 33 (47.1) | 4 (12.1) | 29 (87.9) | 9 (27.3) | 24 (72.7) | 10 (30.3) | 23 (69.7) | 9 (27.3) | 24 (72.7) | 16 (48.5) | 17 (51.5) | 18 (54.5) | 15 (45.5) | 23 (69.7) | 10 (30.3) |
|
Yes | 37 (52.9) | 13 (35.1) | 24 (64.9) | 13 (35.1) | 24 (64.9) | 18 (48.6) | 19 (51.4) | 10 (27.0) | 27 (73.0) | 18 (48.6) | 19 (51.4) | 21 (56.8) | 16 (43.2) | 24 (64.9) | 13 (35.1) |
|
P-value |
| 0.025 |
| 0.479 |
| 0.118 |
| 0.982 |
| 0.989 |
| 0.853 |
| 0.667 |
|
| Menopausal
status |
|
|
Premenopausal | 30 (42.9) | 10 (33.3) | 20 (66.7) | 7 (23.3) | 23 (76.7) | 6 (20.0) | 24 (80.0) | 6 (20.0) | 24 (80.0) | 10 (33.3) | 20 (66.7) | 15 (50.0) | 15 (50.0) | 20 (66.7) | 10 (33.3) |
|
Postmenopausal | 40 (57.1) | 7 (17.5) | 33 (82.5) | 15 (37.5) | 25 (62.5) | 22 (55.0) | 18 (45.0) | 13 (32.5) | 27 (67.5) | 24 (60.0) | 16 (40.0) | 24 (60.0) | 16 (40.0) | 27 (67.5) | 13 (32.5) |
|
P-value |
| 0.126 |
| 0.206 |
| 0.003 |
| 0.244 |
| 0.027 |
| 0.405 |
| 0.941 |
|
| ER |
|
|
Positive | 45 (64.3) | 12 (26.7) | 33 (73.3) | 11 (24.4) | 34 (75.6) | 16 (35.6) | 29 (64.4) | 12 (26.7) | 33 (73.3) | 20 (44.4) | 25 (55.6) | 19 (42.2) | 26 (57.8) | 31 (68.9) | 14 (31.1) |
|
Negative | 25 (35.7) | 5 (20.0) | 20 (80.0) | 11 (44.0) | 14 (56.0) | 12 (48.0) | 13 (52.0) | 7 (28.0) | 18 (72.0) | 14 (56.0) | 11 (44.0) | 20 (80.0) | 5 (20.0) | 16 (64.0) | 9 (36.0) |
|
P-value |
| 0.533 |
| 0.091 |
| 0.309 |
| 0.904 |
| 0.354 |
| 0.002 |
| 0.676 |
|
| PR |
|
|
Positive | 40 (57.1) | 10 (25.0) | 30 (75.0) | 10 (25.0) | 30 (75.0) | 14 (35.0) | 26 (65.0) | 10 (25.0) | 30 (75.0) | 18 (45.0) | 22 (55.0) | 14 (35.0) | 26 (65.0) | 29 (72.5) | 11 (27.5) |
|
Negative | 30 (42.9) | 7 (23.3) | 23 (76.7) | 12 (40.0) | 18 (60.0) | 14 (46.7) | 16 (53.3) | 9 (30.0) | 21 (70.0) | 16 (53.3) | 14 (46.7) | 25 (83.3) | 5 (16.7) | 18 (60.0) | 12 (40.0) |
|
P-value |
| 0.872 |
| 0.181 |
| 0.324 |
| 0.642 |
| 0.490 |
| <0.001 |
| 0.271 |
|
| HER2 |
|
|
Positive | 36 (51.4) | 10 (27.8) | 26 (72.2) | 11 (30.6) | 25 (69.4) | 14 (38.9) | 22 (61.1) | 11 (30.6) | 25 (69.4) | 20 (55.6) | 16 (44.4) | 22 (61.1) | 14 (38.9) | 23 (63.9) | 13 (36.1) |
|
Negative | 34 (48.6) | 7 (20.6) | 27 (79.4) | 11 (32.4) | 23 (67.6) | 14 (41.2) | 20 (58.8) | 8 (23.5) | 26 (76.5) | 14 (41.2) | 20 (58.8) | 17 (50.0) | 17 (50.0) | 24 (70.6) | 10 (29.4) |
|
P-value |
| 0.483 |
| 0.871 |
| 0.845 |
| 0.509 |
| 0.229 |
| 0.350 |
| 0.551 |
|
| TNBC |
|
| No | 12 (17.1) | 2 (16.7) | 10 (83.3) | 6 (50.0) | 6 (50.0) | 4 (33.3) | 8 (66.7) | 2 (16.7) | 10 (83.3) | 7 (58.3) | 5 (41.7) | 9 (75.0) | 3 (25.0) | 10 (83.3) | 2 (16.7) |
|
Yes | 58 (82.9) | 15 (25.9) | 43 (74.1) | 16 (27.6) | 42 (72.4) | 24 (41.4) | 34 (58.6) | 17 (29.3) | 41 (70.7) | 27 (46.6) | 31 (53.4) | 30 (51.7) | 28 (48.3) | 37 (63.8) | 21 (36.2) |
|
P-value |
| 0.759 |
| 0.238 |
| 0.846 |
| 0.589 |
| 0.457 |
| 0.140 |
| 0.330 |
|
| P53 |
|
|
Positive | 38 (54.3) | 9 (23.7) | 29 (76.3) | 12 (31.6) | 26 (68.4) | 10 (26.3) | 28 (73.7) | 9 (23.7) | 29 (76.3) | 15 (39.5) | 23 (60.5) | 26 (68.4) | 12 (31.6) | 23 (60.5) | 15 (39.5) |
|
Negative | 32 (45.7) | 8 (25.0) | 24 (75.0) | 10 (31.2) | 22 (68.8) | 18 (56.2) | 14 (43.8) | 10 (31.2) | 22 (68.8) | 19 (59.4) | 13 (40.6) | 13 (40.6) | 19 (59.4) | 24 (75.0) | 8 (25.0) |
|
P-value |
| 0.898 |
| 0.976 |
| 0.011 |
| 0.478 |
| 0.097 |
| 0.020 |
| 0.199 |
|
Association between gene methylation
and protein expression
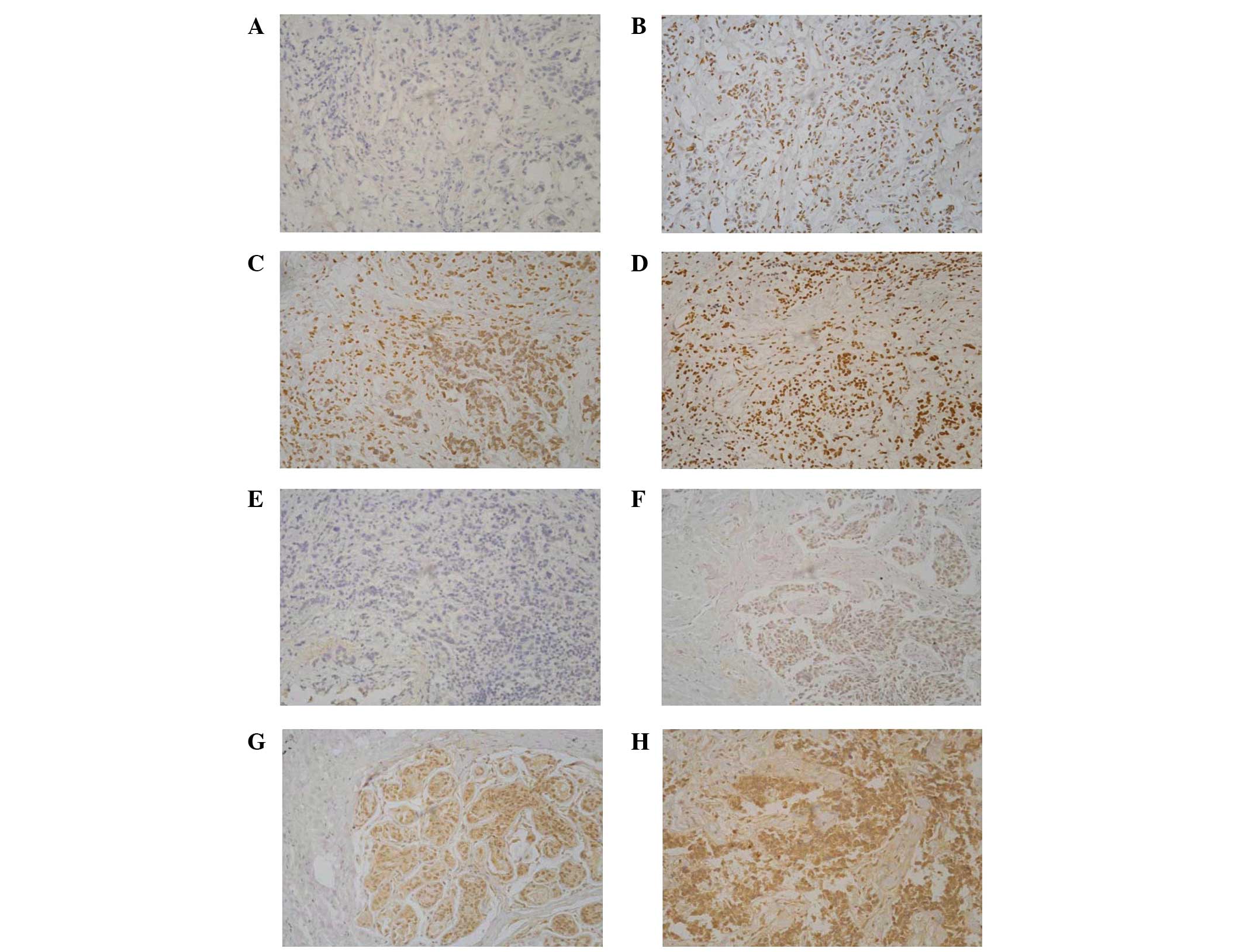
Immunohistochemical analysis was performed to assess
the expression of BRCA1 and GSTP1. With the increase
of methylation frequency, protein expression decreased
significantly (P<0.05; Table
VII). Immunohistochemical staining results together with the
promoter methylation status of BRCA1 and GSTP1 are
shown in Fig. 3.
 | Table VII.Association between methylation and
protein expression. |
Table VII.
Association between methylation and
protein expression.
|
| BRCA1, n
(%) | GSTP1, n
(%) |
|---|
|
|
|
|
|---|
| Gene
expression | M | U | M | U |
|---|
| Negative | 0 (0.0) | 0 (0.0) | 9 (100.0) | 0 (0.0) |
| Weak | 14 (70.0) | 6 (30.0) | 9 (64.3) | 5 (35.7) |
| Moderate | 3 (30.0) | 7 (70.0) | 4 (50.0) | 4 (50.0) |
| Strong | 0 (0.0) | 2 (100.0) | 0 (0.0) | 1 (100.0) |
| P-value for
trend | 0.011 | 0.008 |
Discussion
Tumor biomarker tests are critical to the
implementation of personalized medicine for patients at risk for or
affected by BC. Newly developed genome-wide methods have revealed
multiple epigenetic alterations that contribute to the
carcinogenesis of BC (29). In the
present study, seven cancer-related genes were selected and their
methylation status was compared between 70 patients with sporadic
BC and 20 controls with BBD. The methylation frequencies of these
candidate genes were consistent with previous published articles
(14,23,30–33). As
expected, hypermethylation of these cancer-related genes was more
frequent in cancer tissues as compared with BBD. Moreover,
immunohistochemical analysis demonstrated that a significant
reduction of gene expression is related to promoter
methylation.
BRCA1, which is a typical tumor suppressor
gene, contributes to the regulation of transcriptional activation,
DNA repair, apoptosis, cell-cycle checkpoint control and
chromosomal remodeling (28). A
previous meta-analysis has provided evidence that BRCA1
methylation is associated with the poor survival of patients with
BC (34). The present study reported
that hypermethylation of the BRCA1 gene promoter was present
in 24.3% of patients with BC, which was significantly associated
with distant metastasis. This result led us to hypothesize that
hypermethylation of the BRCA1 gene promoter seemed to confer
an advantage for tumors cells invasion, and may be used as a
biomarker of advanced BC. The present study results demonstrated
that BRCA1 expression was significantly decreased in
patients with BRCA1 hypermethylation, which was consistent
with the results reported by Shilpa et al (35). No methylation of BRCA1 gene was
detected in BBD patients.
GSTP1, which has an important role in the
detoxification of toxic substances, is a phase II metabolic enzyme.
The silencing of phase II metabolic enzymes by promoter methylation
has been suggested to be implicated in the pathogenesis of BC
(36). In the present study, the
frequency of GSTP1 methylation in BBD tissues was 0%.
GSTP1 protein expression was found to be absent or markedly
decreased in the majority of the GSTP1 methylated tumors,
suggesting that epigenetic gene silencing in these tumors may
interfere directly with the binding of sequence-specific
transcription factors that would otherwise promote gene expression
(37).
P16INK4A is a major target in
human carcinogenesis. It is epigenetically silenced in various
human tumors and the downregulation of P16INK4A
protein has been reported in multiple cancers (38). In the present study,
P16INK4A hypermethylation was found to be
associated with patient's age at the diagnosis and menopausal
status. This suggested that loss of P16INK4A
expression through aberrant promoter methylation may occur more
frequently in old women with BC (39). Moreover, it was observed that
P53 expression was associated with
P16INK4A methylation status. A previous study has
revealed that P16INK4A gene activity inversely
modulated p53 status and function in primary human mammary
epithelial cells (40). Reduced
levels of P16INK4A protein stabilize P53
protein through the inhibition of proteolytic degradation (40). Inactivation of
P16INK4A/retinoblastoma and P53/P21
pathways via hypermethylation has been linked to critical telomere
shortening, leading to genome instability and ultimately to BC
formation (41). This may partly
explain the association between P16INK4A
methylation and P53 expression.
MGMT catalyzes the transfer of the methyl
group from O6-methylguanine to a cysteine residue of its active
site (42). Tumors with low levels of
protein expression due to the epigenetic silencing of MGMT
in the promoter region have previously been examined (42). Consistent with the present findings,
earlier studies have reported the frequency of MGMT
methylation ranging from 22 to 32% (43).
CCND2 belongs to a family of D-type cyclins
(44). Previous studies revealed that
high methylation levels of CCND2 caused deregulation of the
G1/S checkpoint, and affected clinicopathologic features of tumor
aggressiveness in BC (44). A study
by Pu et al (45) reported
that methylation of CCND2 (71%) was higher in invasive BCs,
which was consistent with the findings of the present study.
PTEN was the first recognized tumor
suppressor with lipid phosphatase activity (46). Zhang et al (46) reported that PTEN
hypermethylation was detected in 31.1% of BC cases, which was lower
than the present results (48.6%). Notably, the present study also
found that PTEN methylation was more frequent in
postmenopausal patients.
Retinoic acid, which has three subtypes (α, β and
γ), induces growth inhibition and apoptosis by regulating gene
expression through its nuclear receptors (47). The human RARβ gene has four
isoforms (β1, β2, β3 and β4), with the β2 isoform being the most
abundant. The protein encoded by RARβ2 functions in
the inhibition of proliferation, apoptosis and senescence (48). Lee et al (49) demonstrated that RARβ2
and the P53 tumor suppressor gene inhibited oncogene-induced
focus formation. Expression of HER2, ER and PR
proteins are considered to be predictive markers for hormone
therapy response in BC (20). The
present study found that RARβ2 was more frequently
hypermethylated in ER-negative, PR-negative and
P53 positive cancer patients, suggesting an interlink
between cancer-related genes.
Studies testing the ability of promoter methylation
profiles to distinguish benign and malignant diseases have yielded
conflicting results (17). A study
that included women with invasive BC, in situ BC and benign
breast disease compared with healthy controls found that promoter
methylation of three genes (APC, Ras association domain
family 1 isoform A and DAPK) was detectable in DNA obtained
from in situ lesions and invasive samples at all tumor
stages (50). However, in another
study, researchers found that fibroadenomas had patterns of
methylation that were similar to those seen in BC cases (51). In the present study, hypermethylation
was observed in some specific genes in patients with BBD, which was
lower than that in BC cases, with a significantly different
methylation rate in the genes of BRCA1 and GSTP1.
When BRCA1 and GSTP1 were combined, the specificity
of diagnosing BC reached 100.0%; however, the sensitivity was only
44.3%. To achieve a more reliable gene panel, sensitivity and
specificity were expected be enhanced by the addition of other
genes that are frequently hypermethylated in BCs to the panel and a
larger population.
The present study has some limitations. Firstly, the
study protocol only focused on the aberrant DNA methylation of
candidate genes. Other epigenetic traits, such as histone
post-transcriptional modifications and non-coding RNAs, and genetic
mutations are also critical for the spatio-temporal regulation of
gene expression (52,53). This may limit the diagnostic accuracy
of biomarkers detected in this study. Secondly, due to the
limitation of the MSP technology, only a few of CpG islands in the
promoter region of genes were measured. The frequency of
hypermethylation detected using MSP cannot fully reflect the
methylation status of this gene. Therefore, to acquire a
comprehensive understanding of DNA methylation status, more
accurate and quantitative methods are required.
Promoter hypermethylation of BRCA1, GSTP1,
P16INK4A, MGMT, PTEN, RARβ2 and CCND2 was
frequently observed in BC and associated with various clinical
pathological features. Hypermethylation of BRCA1 and
GSTP1 was more common in cancerous tissues, which indicates
these may be used as promising biomarkers for the diagnosis of
BC.
Acknowledgements
The present study was supported by the National
Natural Science Foundation of China (grant no. 81172268), Key
University Science Research Project of Jiangsu Province (grant no.
12KJA330001), Social Development Project in Jiangsu Province (grant
no. BE2015694), Six Talent Peaks Project in Jiangsu Province (grant
no. 2014-YY-023), and Priority Academic Program Development of
Jiangsu Higher Education Institutions (grant no. PAAD).
References
|
1
|
WHO, . Breast cancer: Prevention and
control. Retrieved October 19, 2016. http://www.who.int/cancer/detection/breastcancer/en/index1.html
|
|
2
|
Radpour R, Barekati Z, Kohler C, Lv Q,
Bürki N, Diesch C, Bitzer J, Zheng H, Schmid S and Zhong XY:
Hypermethylation of tumor suppressor genes involved in critical
regulatory pathways for developing a blood-based test in breast
cancer. PLoS One. 6:e160802011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Swedish Organised Service Screening
Evaluation Group, . Reduction in breast cancer mortality from the
organised service screening with mammography: 2. Validation with
alternative analytic methods. Cancer Epidemiol Biomarkers Prev.
15:52–56. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Nelson HD, O'Meara ES, Kerlikowske K,
Balch S and Miglioretti D: Factors associated with rates of
false-positive and false-negative results from digital mammography
screening: An analysis of registry data. Ann Intern Med.
164:226–235. 2016. View
Article : Google Scholar : PubMed/NCBI
|
|
5
|
Njor SH, Hallas J, Schwartz W, Lynge E and
Pedersen AT: Type of hormone therapy and risk of misclassification
at mammography screening. Menopause. 18:171–177. 2011.PubMed/NCBI
|
|
6
|
Baylin SB, Belinsky SA and Herman JG:
Aberrant methylation of gene promoters in cancer-concepts,
misconcepts, and promise. J Natl Cancer Inst. 92:1460–1461. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Jones PA and Laird PW: Cancer epigenetics
comes of age. Nat Genet. 21:163–167. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Toh Y, Egashira A and Yamamoto M:
Epigenetic alterations and their clinical implications in
esophageal squamous cell carcinoma. Gen Thorac Cardiovasc Surg.
61:262–269. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Fang F, Turcan S, Rimner A, Kaufman A,
Giri D, Morris LG, Shen R, Seshan V, Mo Q, Heguy A, et al: Breast
cancer methylomes establish an epigenomic foundation for
metastasis. Sci Transl Med. 3:75ra252011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Jones PA and Baylin SB: The epigenomics of
cancer. Cell. 128:683–692. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kim D Cheol, Thorat MA, Lee MR, Cho SH,
Vasiljević N, Scibior-Bentkowska D, Wu K, Ahmad AS, Duffy S, Cuzick
JM and Lorincz AT: Quantitative DNA methylation and recurrence of
breast cancer: A study of 30 candidate genes. Cancer Biomark.
11:75–88. 2012.PubMed/NCBI
|
|
12
|
Pang JM, Deb S, Takano EA, Byrne DJ, Jene
N, Boulghourjian A, Holliday A, Millar E, Lee CS, O'Toole SA, et
al: Methylation profiling of ductal carcinoma in situ and its
relationship to histopathological features. Breast Cancer Res.
16:4232014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Brooks JD, Cairns P, Shore RE, Klein CB,
Wirgin I, Afanasyeva Y and Zeleniuch-Jacquotte A: DNA methylation
in pre-diagnostic serum samples of breast cancer cases: Results of
a nested case-control study. Cancer Epidemiol. 34:717–723. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Fackler MJ, McVeigh M, Evron E, Garrett E,
Mehrotra J, Polyak K, Sukumar S and Argani P: DNA methylation of
RASSF1A, HIN-1, RAR-beta, Cyclin D2 and Twist in in situ and
invasive lobular breast carcinoma. Int J Cancer. 107:970–975. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Fackler MJ, McVeigh M, Mehrotra J, Blum
MA, Lange J, Lapides A, Garrett E, Argani P and Sukumar S:
Quantitative multiplex methylation-specific PCR assay for the
detection of promoter hypermethylation in multiple genes in breast
cancer. Cancer Res. 64:4442–4452. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Terry MB, McDonald JA, Wu HC, Eng S and
Santella RM: Epigenetic biomarkers of breast cancer risk: Across
the breast cancer prevention continuum. Adv Exp Med Biol.
882:33–68. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Brooks J, Cairns P and Zeleniuch-Jacquotte
A: Promoter methylation and the detection of breast cancer. Cancer
Causes Control. 20:1539–1550. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Sturgeon SR, Balasubramanian R, Schairer
C, Muss HB, Ziegler RG and Arcaro KF: Detection of promoter
methylation of tumor suppressor genes in serum DNA of breast cancer
cases and benign breast disease controls. Epigenetics. 7:1258–1267.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Singletary SE and Connolly JL: Breast
cancer staging: Working with the sixth edition of the AJCC Cancer
Staging Manual. CA Cancer J Clin. 56:37–47. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Amara K, Trimeche M, Ziadi S, Laatiri A,
Hachana M, Sriha B, Mokni M and Korbi S: Presence of simian virus
40 DNA sequences in diffuse large B-cell lymphomas in Tunisia
correlates with aberrant promoter hypermethylation of multiple
tumor suppressor genes. Int J Cancer. 121:2693–2702. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Baldwin RL, Nemeth E, Tran H, Shvartsman
H, Cass I, Narod S and Karlan BY: BRCA1 promoter region
hypermethylation in ovarian carcinoma: A population-based study.
Cancer Res. 60:5329–5333. 2000.PubMed/NCBI
|
|
22
|
Pongtheerat T, Pakdeethai S, Purisa W,
Chariyalertsak S and Petmitr S: Promoter methylation and genetic
polymorphism of glutathione S-transferase P1 gene (GSTP1) in Thai
breast- cancer patients. Asian Pac J Cancer Prev. 12:2731–2734.
2011.PubMed/NCBI
|
|
23
|
Vallian S, Sedaghat M, Nassiri I and
Frazmand A: Methylation status of p16 INK4A tumor suppressor gene
in Iranian patients with sporadic breast cancer. J Cancer Res Clin
Oncol. 135:991–996. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ye C, Shrubsole MJ, Cai Q, Ness R, Grady
WM, Smalley W, Cai H, Washington K and Zheng W: Promoter
methylation status of the MGMT, hMLH1, and CDKN2Ap16 genes in
non-neoplastic mucosa of patients with and without colorectal
adenomas. Oncol Rep. 16:429–435. 2006.PubMed/NCBI
|
|
25
|
Muñoz J, Inda MM, Lázcoz P, Zazpe I, Fan
X, Alfaro J, Tuñón T, Rey JA and Castresana JS: Promoter
methylation of RASSF1A associates to adult secondary glioblastomas
and pediatric glioblastomas. ISRN Neurol. 2012:5765782012.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Karray-Chouayekh S, Trifa F, Khabir A,
Boujelbane N, Sellami-Boudawara T, Daoud J, Frikha M, Jlidi R,
Gargouri A and Mokdad-Gargouri R: Aberrant methylation of RASSF1A
is associated with poor survival in Tunisian breast cancer
patients. J Cancer Res Clin Oncol. 136:203–210. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Matsubayashi H, Sato N, Fukushima N, Yeo
CJ, Walter KM, Brune K, Sahin F, Hruban RH and Goggins M:
Methylation of cyclin D2 is observed frequently in pancreatic
cancer but is also an age-related phenomenon in gastrointestinal
tissues. Clin Cancer Res. 9:1446–1452. 2003.PubMed/NCBI
|
|
28
|
Bai X, Fu Y, Xue H, Guo K, Song Z, Yu Z,
Jia T, Yan Y, Zhao L, Mi X, et al: BRCA1 promoter hypermethylation
in sporadic epithelial ovarian carcinoma: Association with low
expression of BRCA1, improved survival and co-expression of DNA
methyltransferases. Oncol Lett. 7:1088–1096. 2014.PubMed/NCBI
|
|
29
|
Dworkin AM, Huang TH and Toland AE:
Epigenetic alterations in the breast: Implications for breast
cancer detection, prognosis and treatment. Semin Cancer Biol.
19:165–171. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Chen Y, Zhou J, Xu Y, Li Z, Wen X, Yao L,
Xie Y and Deng D: BRCA1 promoter methylation associated with poor
survival in Chinese patients with sporadic breast cancer. Cancer
Sci. 100:1663–1667. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Shinozaki M, Hoon DS, Giuliano AE, Hansen
NM, Wang HJ, Turner R and Taback B: Distinct hypermethylation
profile of primary breast cancer is associated with sentinel lymph
node metastasis. Clin Cancer Res. 11:2156–2162. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Sharma G, Mirza S, Parshad R, Srivastava
A, Gupta SD, Pandya P and Ralhan R: Clinical significance of
promoter hypermethylation of DNA repair genes in tumor and serum
DNA in invasive ductal breast carcinoma patients. Life Sci.
87:83–91. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Sadeq V, Isar N and Manoochehr T:
Association of sporadic breast cancer with PTENMMAC1TEP1 promoter
hypermethylation. Med Oncol. 28:420–423. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wu L, Wang F, Xu R, Zhang S, Peng X, Feng
Y, Wang J and Lu C: Promoter methylation of BRCA1 in the prognosis
of breast cancer: A meta-analysis. Breast Cancer Res Treat.
142:619–627. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Shilpa V, Bhagat R, Premalata CS, Pallavi
VR, Ramesh G and Krishnamoorthy L: BRCA1 promoter hypermethylation
and protein expression in ovarian carcinoma-an Indian study. Tumour
Biol. 35:4277–4284. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Esteller M, Corn PG, Urena JM, Gabrielson
E, Baylin SB and Herman JG: Inactivation of glutathione
S-transferase P1 gene by promoter hypermethylation in human
neoplasia. Cancer Res. 58:4515–4518. 1998.PubMed/NCBI
|
|
37
|
Ronneberg JA, Tost J, Solvang HK, Alnaes
GI, Johansen FE, Brendeford EM, Yakhini Z, Gut IG, Lønning PE,
Børresen-Dale AL, et al: GSTP1 promoter haplotypes affect DNA
methylation levels and promoter activity in breast carcinomas.
Cancer Res. 68:5562–5571. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Lee JJ, Ko E, Cho J, Park HY, Lee JE, Nam
SJ, Kim DH and Cho EY: Methylation and immunoexpression of p16
(INK4a) tumor suppressor gene in primary breast cancer tissue and
their quantitative p16 (INK4a) hypermethylation in plasma by
real-time PCR. Korean J Pathol. 46:554–561. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Johnson KC, Koestler DC, Cheng C and
Christensen BC: Age-related DNA methylation in normal breast tissue
and its relationship with invasive breast tumor methylation.
Epigenetics. 9:268–275. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Zhang J, Pickering CR, Holst CR, Gauthier
ML and Tlsty TD: p16INK4a modulates p53 in primary human mammary
epithelial cells. Cancer Res. 66:10325–10331. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Radpour R, Barekati Z, Haghighi MM, Kohler
C, Asadollahi R, Torbati PM, Holzgreve W and Zhong XY: Correlation
of telomere length shortening with promoter methylation profile of
p16Rb and p53p21 pathways in breast cancer. Mod Pathol. 23:763–772.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Ingold B, Schraml P, Heppner FL and Moch
H: Homogeneous MGMT immunoreactivity correlates with an
unmethylated MGMT promoter status in brain metastases of various
solid tumors. PLoS One. 4:e47752009. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Asiaf A, Ahmad ST, Malik AA, Aziz SA,
Rasool Z, Masood A and Zargar MA: Protein expression and
methylation of MGMT, a DNA repair gene and their correlation with
clinicopathological parameters in invasive ductal carcinoma of the
breast. Tumour Biol. 36:6485–6496. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Sharma G, Mirza S, Prasad CP, Srivastava
A, Gupta SD and Ralhan R: Promoter hypermethylation of p16INK4A,
p14ARF, CyclinD2 and Slit2 in serum and tumor DNA from breast
cancer patients. Life Sci. 80:1873–1881. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Pu RT, Laitala LE, Alli PM, Fackler MJ,
Sukumar S and Clark DP: Methylation profiling of benign and
malignant breast lesions and its application to cytopathology. Mod
Pathol. 16:1095–1101. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Zhang HY, Liang F, Jia ZL, Song ST and
Jiang ZF: Mutation, methylation and expression in breast cancer
patients. Oncol Lett. 6:161–168. 2013.PubMed/NCBI
|
|
47
|
Kim Y, Jin D, Lee BB, Cho EY, Han J, Shim
YM and Kim DH: RARbeta2 hypermethylation is associated with poor
recurrence-free survival in never-smokers with adenocarcinoma of
the lung. Clinical Epigenetics. 7:322015. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Tao MH, Shields PG, Nie J, Millen A,
Ambrosone CB, Edge SB, Krishnan SS, Marian C, Xie B, Winston J, et
al: DNA hypermethylation and clinicopathological features in breast
cancer: The Western New York exposures and breast cancer (WEB)
study. Breast Cancer Res Treat. 114:559–568. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Lee X, Si SP, Tsou HC and Peacocke M:
Cellular aging and transformation suppression: A role for retinoic
acid receptor beta 2. Exp Cell Res. 218:296–304. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Dulaimi E, Hillinck J, de Caceres I
Ibanez, Al-Saleem T and Cairns P: Tumor suppressor gene promoter
hypermethylation in serum of breast cancer patients. Clin Cancer
Res. 10:6189–6193. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Cowin P, Rowlands TM and Hatsell SJ:
Cadherins and catenins in breast cancer. Curr Opin Cell Biol.
17:499–508. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Falahi F, Sgro A and Blancafort P:
Epigenome engineering in cancer: Fairytale or a realistic path to
the clinic? Front Oncol. 5:222015. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Connolly R and Stearns V: Epigenetics as a
therapeutic target in breast cancer. J Mammary Gland Biol
Neoplasia. 17:191–204. 2012. View Article : Google Scholar : PubMed/NCBI
|