Introduction
Tongue squamous cell carcinoma (TSCC), the most
common type of oral cancer, often leads to malfunctions in speech,
mastication and deglutition (1). TSCC
is well-known for its high rate of proliferation and nodal
metastasis; metastasis is the most reliable adverse prognostic
factor in TSCC patients (2). Despite
being visibly located in the oral cavity, >50% patients at
diagnosis present with advanced stage III or IV according to the
tumor-node metastasis classification of malignant tumors system
(3). Despite improvements in surgery,
radiotherapy and chemotherapy, the 5-year survival rate for
patients with TSCC remains poor, mainly due to regional recurrence
and lymph node metastasis (4,5). An accumulating number of studies suggest
that TSCC arises as a result of oncogene activation or tumor
suppressor gene inactivation (6–8). However,
the detailed molecular mechanism of TSCC remain unknown (5). Understanding the molecular pathways of
TSCC carcinogenesis and development may be useful for improving
diagnosis, treatment and prevention of the disease.
Altered expression of microRNA (miRNA) has been
found in numerous types of cancer (9). miRNA is a type of small,
single-stranded, non-coding RNA of between 18 and 25 nucleotides in
length that performs an important role in the posttranscriptional
regulation of gene expression (10).
In total, >1,800 types of miRNA have been identified in miRBase
version 20.0, and 1/3 of the genes in the human genome are
regulated by miRNA (11,12). miRNA regulates target gene expression
post-transcriptionally via incomplete base pairing with the target
mRNA (13). Furthermore, miRNA
performs an important role in numerous biological processes
including development, differentiation, proliferation, apoptosis,
angiogenesis and metabolism. Mutations of miRNA have consequently
been suggested to serve an important role in carcinogenesis
(14). In addition, miRNA can
function as either tumor suppressors or oncogenes, depending on
whether oncogenes or tumor suppressor genes are targeted (15). Thus, identifying miRNA targets is
critical to understand the function of miRNA in tumorigenesis and
progression. The present study also suggests that miRNAs may be a
target for cancer therapy.
The expression and function of (miRNA-149) miR-149
have been investigated in a number of types of cancer. This study
was aimed to investigate the expression, biological functions and
molecular mechanisms of miR-149 in TSCC. The present study found
that the expression of miR-149 was decreased in TSCC tissues and
cell lines compared with matched normal tissue and normal gingival
epithelial cells, respectively. Additionally, the present study
demonstrated that miR-149 suppressed cell proliferation, migration
and invasion by directly targeting specificity protein 1 (SP1). The
present results improve the understanding of the mechanisms of TSCC
carcinogenesis and progression, and identify new targets that may
be used for the development of novel treatments of TSCC.
Materials and methods
Clinical specimens
TSCC tissue and matched normal adjacent tissue (NAT)
for quantitative polymerase chain reaction (qPCR) were obtained
from 62 patients who had undergone primary surgical treatment of
oral tongue carcinoma at Nanfang Hospital (Guangzhou, China). None
of the patients received treatment prior to the excision surgery.
The tissues were flash frozen in liquid nitrogen and stored at
−80°C until use. The present study was approved by the Protection
of Human Subjects Committee of Nanfang Hospital (Guangzhou, China).
Written informed consent was also acquired from all TSCC
patients.
Cell culture
TSCC Tca8113 and CAL-27 cell lines and normal
gingival epithelial cells were purchased from the American Type
Culture Collection (Manassas, VA, USA). The Tca8113 and CAL-27 cell
lines were cultured in RPMI 1640 medium (Gibco; Thermo Fisher
Scientific, Inc., Waltham, MA, USA), while normal gingival
epithelial cells were maintained in minimum essential media (Gibco;
Thermo Fisher Scientific, Inc.). All media were supplemented with
10% (v/v) fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific,
Inc.), 100 U/ml penicillin and 100 U/ml streptomycin. All cell
lines were cultured at 37°C in a humidified air atmosphere
containing 5% CO2.
Cell transfection
miR-149 mimics and miRNA mimics negative control
(NC), obtained from Shanghai GenePharma Co., Ltd., (Shanghai,
China), were used for the upregulation of miR-149 activity in
cells. Transfection was completed using Lipofectamine 2000
(Invitrogen, Thermo Fisher Scientific, Inc.), according to the
manufacturer's protocol.
RNA isolation, reverse transcription
and qPCR
Total RNA was extracted from the TSCC tissue and NAT
cells and the Tca8113, CAL-27 and normal gingival epithelial cells
using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.)
according to the manufacturer's protocol. For complementary DNA
synthesis, 1 µg RNA was mixed with 500 ng oligo-deoxythymine
(Promega Corporation, Madison, WI, USA) or microRNA specific
primers (Invitrogen; Thermo Fisher Scientific, Inc.). The
GoScipt/ImProm-II reverse transcription system (Promega
Corporation) was then used to perform reverse transcription on the
samples. The temperature protocol was as follows: 95°C for 2 min;
20 cycles of 94°C for 1 min; 55°C for 1 min and 72°C for 2 min; and
72°C for 5 min. qPCR was performed using an Applied Biosystems 7500
Real-time PCR system (Thermo Fisher Scientific, Inc.) and an SYBR
premix Ex Taq kit (Takara, Biotechnology Co., Ltd., Dalian, China)
following the manufacturer's protocol. All samples were amplified
in triplicate. The levels of miR-149 were normalized using U6 small
nuclear RNA as the endogenous reference gene. The primers were
obtained from Guangzhou RiboBio Co., Ltd. (Guangzhou, China), and
sequences were as follows: miR-149 forward,
5′-TCTGGCTCCGTGTCTTCACTCCC-3′ and reverse,
5′-AGTGGTTGTTCTGCTCTCTGTGTC-3′; U6 forward,
5′-CTCGCTTCGGCAGCACATATACT-3′ and reverse,
5′-ACGCTTCACGAATTTGCGTGTC-3′. Relative expression fold changes were
calculated using the 2−ΔΔCq method (16).
Cell proliferation assay
To explore the effect of miR-149 on cell
proliferation, an MTT assay was used. Tca8113 and CAL-27 cells
transfected with miR-149 or negative control (NC) were seeded in
96-well plates at a density of 3,000 cells/well. The MTT assay was
performed by incubating the cells with 20 µl MTT (5 mg/ml;
Sigma-Aldrich, Merck Millipore, Darmstadt, Germany). Subsequent to
incubation for 4 h at 37°C, the formazan precipitates were
dissolved in 200 µl dimethyl sulfoxide. Absorbance at 490 nm was
determined using an ELISA reader (Bio-Rad Laboratories, Inc.,
Hercules, CA, USA). All experiments were repeated at least 3
times.
Cell migration and invasion
assays
The migration and invasion of the TSCC cell lines
was examined using Transwell chambers (8 µm; EMD Millipore,
Billerica, MA, USA). The cell migration assay was performed with
uncoated Matrigel (BD Biosciences, San Jose, CA, USA) whereas the
cell invasion assays were performed with coated Matrigel (BD
Biosciences). A total of 5×104 Tca8113 and CAL-27 cells
transfected with miR-149 or NC in 200 µl serum-free RPMI 1640
medium (Gibco; Thermo Fisher Scientific, Inc.) were seeded into the
upper chamber. A volume of 500 µl RPMI 1640 medium containing 10%
FBS was then added to the lower chamber as a chemoattractant. The
cells were then incubated for 12 h, for the migration assay, and 24
h for the invasion assay. The cells remaining on the upper surface
of the membranes were scraped off using cotton swabs and the
membranes were fixed with 100% methanol (Shanghai Macklin
Biochemical Co., Ltd, Shanghai, China) for 10 min and stained in
0.5% crystal violet (Beyotime Institute of Biotechnology, Haimen,
China). The membranes were counted under an inverted microscope
(CKX41; Olympus Corporation, Tokyo, Japan) to calculate their
relative numbers. Each experiment was repeated at least 3
times.
Western blotting
The TSCC cell lines were plated in 6-well plates. A
total of 72 h subsequent to transfection with miR-149 or NC, the
cells were washed and lysed in cold radioimmunoprecipitation assay
lysis buffer (Beyotime Institute of Biotechnology). Subsequent to
centrifugation at 24,150 × g for 10 min at 4°C, the protein
concentration was determined using a bicinchoninic acid Protein
Assay kit (Beyotime Institute of Biotechnology). Equal amounts of
protein were then separated by 10% SDS-PAGE and transferred to
polyvinylidene fluoride membranes (Beyotime Institute of
Biotechnology). The membranes were blocked with 5% skimmed milk at
room temperature for 2 h and incubated with rabbit anti-human SP1
antibody (dilution, 1: 1,000; cat. no., 5931; Cell Signaling
Technology, Inc., Danvers, MA, USA) according to the protocol of
the manufacturer at 4°C overnight. The membranes were washed and
then incubated with the corresponding horseradish
peroxidase-conjugated secondary antibody (dilution, 1:1,000; cat.
no., 7074; Cell Signaling Technology, Inc., Danvers, MA, USA) for 1
h. The protein bands were visualized with an enhanced
chemiluminescence kit (Pierce, Thermo Fisher Scientific, Inc.) and
analyzed using Quantity One software (version 4.62; Bio-Rad
Laboratories, Inc.).
Luciferase assay
The human TSCC cell lines were plated in a 12-well
plate at ~90% confluence and transfected with a reporter plasmid,
miR-49 mimic or NC by Lipofectamine 2000 (Thermo Fisher Scientific,
Inc.) according to the protocol of the manufacturer. The activity
of Photinus and Renilla luciferase in cell lysates was determined
with the Dual-Luciferase Reporter Assay System (Promega
Corporation) following 48 h transfection. The Photinus luciferase
activity was normalized to the Renilla luciferase activity for each
transfected well. All the experiments were performed in
triplicate.
Statistical analysis
Data were presented as the mean ± standard deviation
and compared using SPSS 13.0 software (SPSS, Inc., Chicago, IL,
USA). P<0.05 was considered to indicate a statistically
significant difference.
Results
miR-149 expression in TSCC tissues and
cell lines
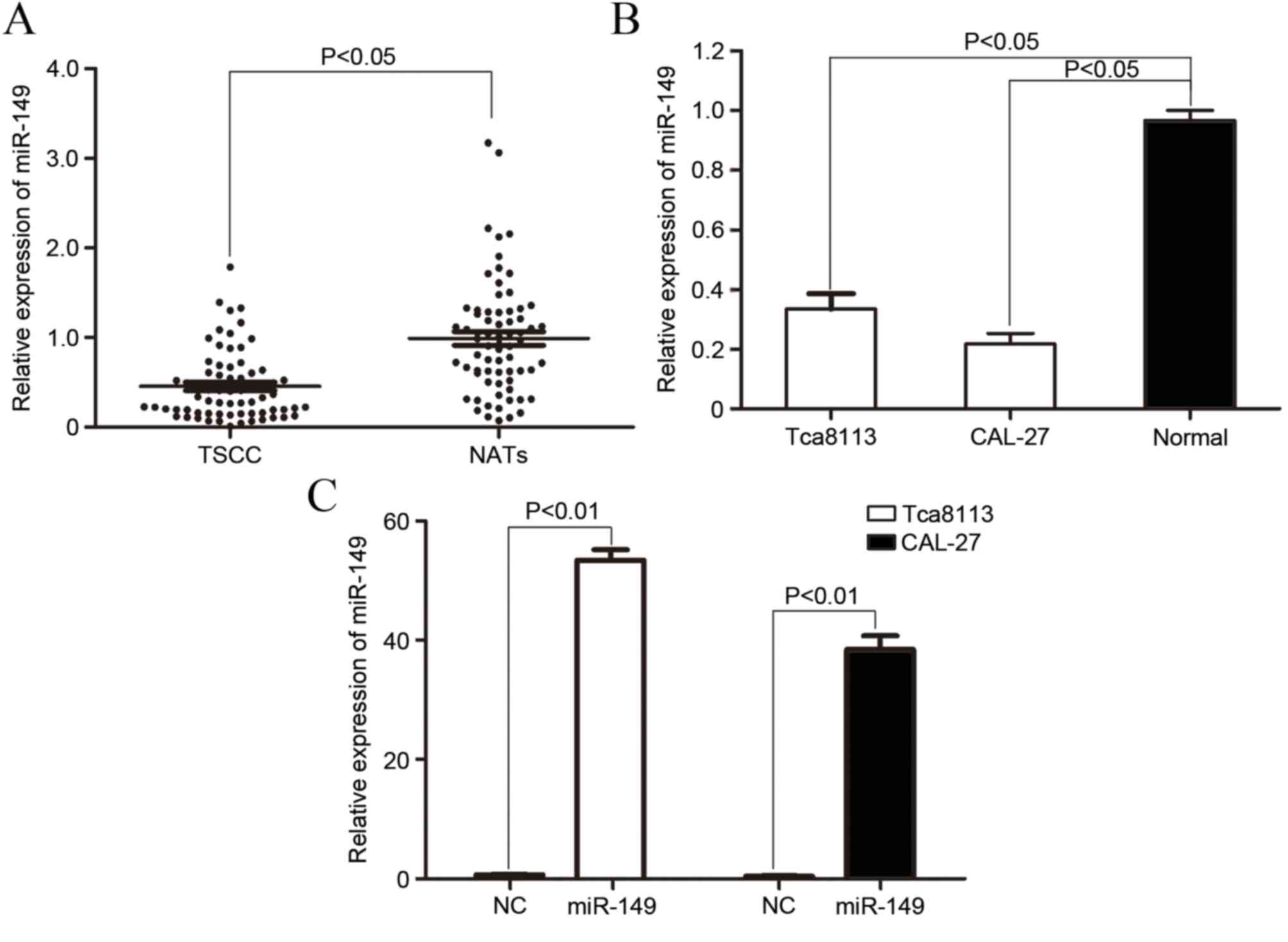
To explore the role of miR-149 in TSCC
carcinogenesis and progression, the expression of miR-149 in TSCC
tissue and NAT was analyzed by qPCR. As demonstrated in Fig. 1A, the expression of miR-149 was
significantly downregulated in TSCC tissues compared with NAT
(P=0.017).
The expression level of miR-149 in TSCC cell lines
and normal gingival epithe-lial cells was also determined. As shown
in Fig. 1B, the miR-149 expression
level significantly decreased in Tca8113 (P=0.01) and CAL-27
(P=0.005) cells compared with normal gingival epithelial cells. The
aforementioned results indicated that miR-149 may serve a role in
TSCC carcinogenesis and progression.
To explore the function of miR-149 on TSCC cells,
the present study transfected the miR-149 mimic into Tca8113 and
CAL-27 cells. As demonstrated in Fig.
1C, the increased level of expression of miR-149 in Tca8113
(P=0.0000017) and CAL-27 (P=0.0000040) cells was confirmed by
qPCR.
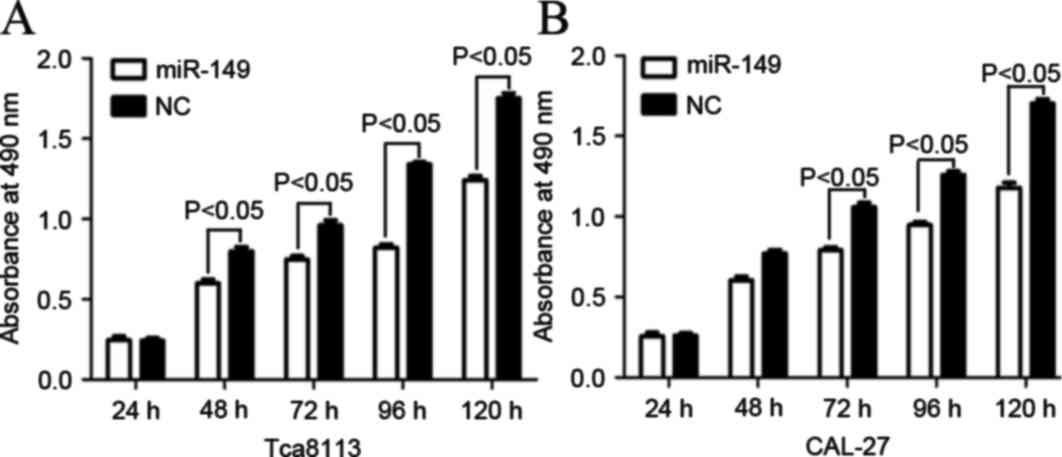
miR-149 inhibited cell proliferation
in Tca8113 and CAL-27 cells
The effect of miR-149 on cell proliferation in
Tca8113 and CAL-27 cells was determined by MTT assay. As shown in
Fig. 2, the level of absorbance in
Tca8113 and CAL-27 cells transfected with miR-149 significantly
decreased compared with cells transfected with NC (P=0.022 for
Tca8113; P=0.018 for CAL-27). The aforementioned results verified
that miR-149 inhibited proliferation in Tca8113 and CAL-27 cell
lines.
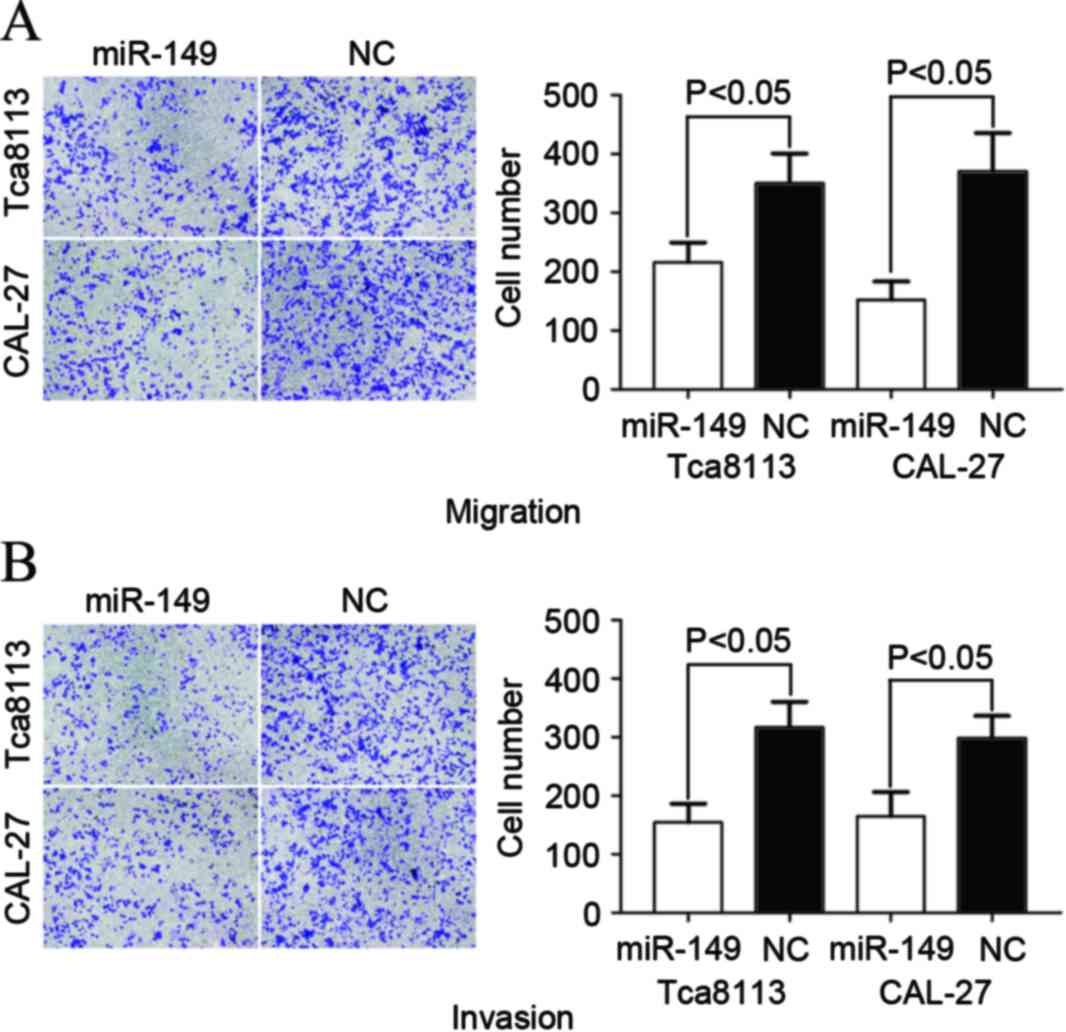
miR-149 suppressed cell migration and
invasion in Tca8113 and CAL-27 cells
A Transwell apparatus assay was performed to explore
the effect of miR-149 on cell migration and invasion. As
demonstrated in Fig. 3, the levels of
migration and invasion of Tca8113 (P=0.034 for migration; P=0.025
for invasion) and CAL-27 (P=0.023 for migration; P=0.031 for
invasion) cells transfected with miR-149 significantly decreased
compared with cells transfected with NC. The aforementioned results
indicated that miR-149 inhibited cell migration and invasion
ability in TSCC cell lines.
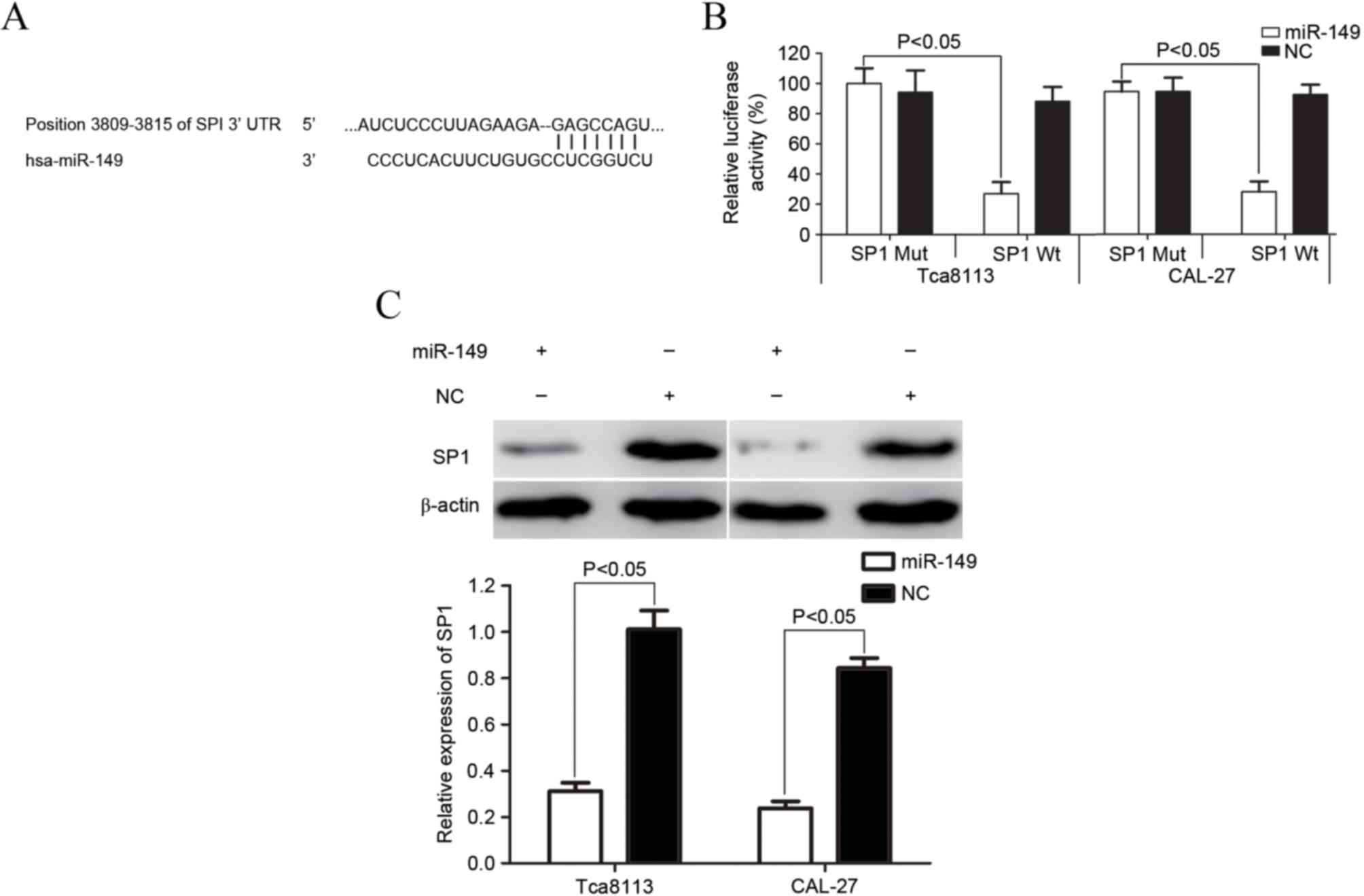
SP1 was a direct target gene of
miR-149 in vitro
To predict the target gene of miR-149,
bioinformatics software (TargetScan 7.1) was used (17). As demonstrated in Fig. 4A, SP1 was verified to be a direct
target gene of miR-149. A luciferase assay was then performed to
investigate whether miR-149 directly targets SP1. As shown in
Fig. 4B, miR-149 significantly
inhibited SP1 wild-type, but not SP1 mutant, luciferase activity in
Tca8113 and CAL-27 cells (P=0.019 for Tca8113; P=0.015 for
CAL-27).
Western blotting was also performed to explore
whether the expression of SP1 was downregulated at the protein
level subsequent to transfection with miR-149 in Tca8113 and CAL-27
cells. As demonstrated in Fig. 4C,
the expression of SP1 significantly decreased in Tca8113 and CAL-27
cells subsequent to transfection with miR-149 (P=0.024 for Tca8113;
P=0.012 for CAL-27). SP1 may therefore be a direct target gene of
miR-149 in vitro.
Discussion
This study demonstrated that the expression levels
of certain types of miRNA decrease in TSCC, and that miRNA may
function as a negative regulator of oncogenes or tumor suppressors
in carcinogenesis and cancer progression. miRNA is thereby a
potential diagnostic and prognostic marker of TSCC with therapeutic
potential (18–20). Previous studies revealed that miR-149
was downregulated in colorectal cancer, breast cancer, gastric
cancer, glioma, melanoma and non-small cell lung cancer (21–26). This
study investigated the expression, biological functions and
molecular mechanisms of miR-149 in TSCC. The present study revealed
that miR-149 was significantly downregulated in TSCC tissue and
cell lines. It also suggested that miR-149 may serve an important
role in TSCC carcinogenesis and development.
An accumulating number of studies indicate that
miR-149 functions as a tumor suppressor in numerous types of human
cancer. For example, in colorectal cancer, miR-149 inhibited cell
migration and invasion by targeting Forkhead Box M1 (21). Chan et al (22) reported that miR-149 targeted
G-protein-coupled receptor kinase-interacting protein 1 to suppress
breast cancer cell migration, invasion and metastasis. In addition,
miR-149 was reported to decrease glioma cell growth and invasion
through targeting protein kinase B signaling (24). Furthermore, Wang et al
(23) found that miR-149 suppressed
cell proliferation and cell cycle progression via the blockade of
zinc finger and BTB domain containing 2 in human gastric cancer. At
present, little is known with respect to the role of miR-149 in
TSCC. The present study demonstrated that the upregulation of
miR-149 inhibited the proliferation, migration and invasion of TSCC
cells. The present study expanded the number of known functions of
miR-149 in cancer.
As miRNA functions by targeting mRNA, the
identification of miR-149 target genes may contribute to the
understanding of the potential role of miRNA in carcinogenesis and
tumor development. In the present study, an important molecular
link between miR-149 and SP1 was observed. Firstly, bioinformatics
software predicted that SP1 possessed a miR-149-targeted seed
sequence, suggesting that SP1 may be a putative target of miR-149.
Secondly, the luciferase assay showed that miR-149 directly
targeted SP1 3′-untranslated region. Finally, western blot analysis
revealed that the upregulation of miR-149 suppressed the expression
of SP1 at a protein level. The aforementioned findings suggested
that miR-149 served a tumor suppressor role in TSCC carcinogenesis
and progression through the direct targeting of SP1.
SP1, a sequence-specific DNA-binding protein, maps
to 12q13.1 and encodes a protein of 785 amino acids (27). It was the first transcription factor
identified and characterized. SP1 is widely expressed in all
mammalian tissues and performs a number of important functions in
normal tissue development (28). The
aberrant expression of SP1 has been found in numbers types of human
cancer, and the involvement of SP1 in carcinogenesis and cancer
progression has also been verified (29–31). An
increasing number of studies suggest that SP1 regulates a variety
of biological functions, including cell survival, proliferation,
differentiation, migration and invasion (32–34).
Additionally, several compounds with anti-tumor effects, which
target SP1 have been developed or are in development, and some of
them are currently used clinically for cancer treatment (35–38).
Therefore, SP1 has been suggested to be a novel target for cancer
therapy due to the cancer-associated functions of the protein.
SP1 has been found to be regulated by multiple types
of miRNA in a variety of types of cancer, including TSCC. miR-29b
and miR-375 function as tumor suppressors in TSCC through targeting
SP1 (39,40). miR-145, miR-133a, miR-133b, miR-22 and
miR-335 serve an important role in the biology of gastric cancer by
regulating SP1 directly (41–43). miR-145 was also found to regulate SP1
in ovarian cancer cells sensitized to paclitaxel (44). In esophageal carcinoma, Wang et
al (45) found that ectopic
miR-429 suppressed cell invasion and induced cell apoptosis via the
blockade of SP1. Zhang et al (46) demonstrated that miR-377 decreased cell
growth and invasion ability by inhibiting SP1. In hepatocellular
carcinoma, the restoration of miR-1188 inhibited cell proliferation
and migration, and enhanced cell apoptosis through the
down-regulation of SP1 (47). In the
present study, the upregulation of miR-149 in TSCC cell lines
revealed that miR-149 inhibited cell proliferation, migration and
invasion via the blockade of SP1. miR-149 may therefore act as a
regulator of SP1.
In conclusion, the present study revealed that
miR149 was significantly downregulated in TSCC tissue and cell
lines. The present study also observed that miR-149 contributed to
cell proliferation, migration and invasion by directly targeting
SP1 in TSCC. The identified candidate target gene of miR-149 may
provide an understanding of potential carcinogenic mechanisms in
TSCC. The findings of the present study have therapeutic
implications and may be exploited for the future treatment of
TSCC.
References
|
1
|
Yu ZW, Zhong LP, Ji T, Zhang P, Chen WT
and Zhang CP: MicroRNAs contribute to the chemoresistance of
cisplatin in tongue squamous cell carcinoma lines. Oral Oncol.
46:317–322. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Yuen PW, Lam KY, Chan AC, Wei WI and Lam
LK: Clinicopathological analysis of local spread of carcinoma of
the tongue. Am J Surg. 175:242–244. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Po Wing Yuen A, Lam KY, Lam LK, Ho CM,
Wong A, Chow TL, Yuen WF and Wei WI: Prognostic factors of
clinically stage I and II oral tongue carcinoma-A comparative study
of stage, thickness, shape, growth pattern, invasive front
malignancy grading, Martinez-Gimeno score, and pathologic features.
Head Neck. 24:513–520. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Sano D and Myers JN: Metastasis of
squamous cell carcinoma of the oral tongue. Cancer Metastasis Rev.
26:645–662. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Song KB, Liu WJ and Jia SS: miR-219
inhibits the growth and metastasis of TSCC cells by targeting
PRKCI. Int J Clin Exp Med. 7:2957–2965. 2014.PubMed/NCBI
|
|
6
|
Squarize CH, Castilho RM, Abrahao AC,
Molinolo A, Lingen MW and Gutkind JS: PTEN deficiency contributes
to the development and progression of head and neck cancer.
Neoplasia. 15:461–471. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Knopf A, Lempart J, Bas M, SlottaHuspenina
J, Mansour N and Fritsche MK: Oncogenes and tumor suppressor genes
in squamous cell carcinoma of the tongue in young patients.
Oncotarget. 6:3443–3451. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Regezi JA, Dekker NP, McMillan A,
RamirezAmador V, MenesesGarcia A, Ruiz-Godoy Rivera LM, Chrysomali
E and Ng IO: p53, p21, Rb, and MDM2 proteins in tongue carcinoma
from patients <35 versus >75 years. Oral Oncol. 35:379–383.
1999. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Calin GA and Croce CM: MicroRNA signatures
in human cancers. Nat Rev Cancer. 6:857–866. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lewis BP, Burge CB and Bartel DP:
Conserved seed pairing, often flanked by adenosines, indicates that
thousands of human genes are microRNA targets. Cell. 120:15–20.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kozomara A and Griffiths-Jones S: miRBase:
Integrating microRNA annotation and deep-sequencing data. Nucleic
Acids Res. 39:(Database Issue). D152–D157. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Shukla GC, Singh J and Barik S: MicroRNAs:
Processing, Maturation, Target Recognition and Regulatory
functions. Mol Cell Pharmacol. 3:83–92. 2011.PubMed/NCBI
|
|
14
|
Ryan BM, Robles AI and Harris CC: Genetic
variation in microRNA networks: The implications for cancer
research. Nat Rev Cancer. 10:389–402. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Cimmino A, Calin GA, Fabbri M, Iorio MV,
Ferracin M, Shimizu M, Wojcik SE, Aqeilan RI, Zupo S, Dono M, et
al: miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl
Acad Sci USA. 102:13944–13949. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Agarwal V, Bell GW, Nam JW and Bartel DP:
Predicting effective microRNA target sites in mammalian mRNAs.
Elife. 4:10.7554/eLife.05005. 2015. View Article : Google Scholar
|
|
18
|
Wong TS, Liu XB, Wong BY, Ng RW, Yuen AP
and Wei WI: Mature miR-184 as Potential Oncogenic microRNA of
Squamous Cell Carcinoma of Tongue. Clin Cancer Res. 14:2588–2592.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Jiang L, Liu X, Chen Z, Jin Y, Heidbreder
CE, Kolokythas A, Wang A, Dai Y and Zhou X: MicroRNA-7 targets
IGF1R (insulin-like growth factor 1 receptor) in tongue squamous
cell carcinoma cells. Biochem J. 432:199–205. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Liu X, Yu J, Jiang L, Wang A, Shi F, Ye H
and Zhou X: MicroRNA-222 regulates cell invasion by targeting
matrix metalloproteinase 1 (MMP1) and manganese superoxide
dismutase 2 (SOD2) in tongue squamous cell carcinoma cell lines.
Cancer Genomics Proteomics. 6:131–139. 2009.PubMed/NCBI
|
|
21
|
Xu K, Liu X, Mao X, Xue L, Wang R, Chen L
and Chu X: MicroRNA-149 suppresses colorectal cancer cell migration
and invasion by directly targeting forkhead box transcription
factor FOXM1. Cell Physiol Biochem. 35:499–515. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Chan SH, Huang WC, Chang JW, Chang KJ, Kuo
WH, Wang MY, Lin KY, Uen YH, Hou MF, Lin CM, et al: MicroRNA-149
targets GIT1 to suppress integrin signaling and breast cancer
metastasis. Oncogene. 33:4496–4507. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wang Y, Zheng X, Zhang Z, Zhou J, Zhao G,
Yang J, Xia L, Wang R, Cai X, Hu H, et al: MicroRNA-149 inhibits
proliferation and cell cycle progression through the targeting of
ZBTB2 in human gastric cancer. PLoS One. 7:e416932012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Pan SJ, Zhan SK, Pei BG, Sun QF, Bian LG
and Sun BM: MicroRNA-149 inhibits proliferation and invasion of
glioma cells via blockade of AKT1 signaling. Int J Immunopathol
Pharmacol. 25:871–881. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Jin L, Hu WL, Jiang CC, Wang JX, Han CC,
Chu P, Zhang LJ, Thorne RF, Wilmott J, Scolyer RA, et al:
MicroRNA-149*, a p53-responsive microRNA, functions as an oncogenic
regulator in human melanoma. Proc Natl Acad Sci USA.
108:15840–15845. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ke Y, Zhao W, Xiong J and Cao R: miR-149
Inhibits Non-small-cell lung cancer cells EMT by targeting FOXM1.
Biochem Res Int. 2013:5067312013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Chang WC and Hung JJ: Functional role of
post-translational modifications of Sp1 in tumorigenesis. J Biomed
Sci. 19:942012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Li J, Zou WX and Chang KS: Inhibition of
Sp1 functions by its sequestration into PML nuclear bodies. PLoS
One. 9:e944502014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Yue L, Li L, Liu F, Hu N, Zhang W, Bai X,
Li Y, Zhang Y, Fu L, Zhang X and Ye L: The oncoprotein HBXIP
activates transcriptional coregulatory protein LMO4 via Sp1 to
promote proliferation of breast cancer cells. Carcinogenesis.
34:927–935. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Yin P, Zhao C, Li Z, Mei C, Yao W, Liu Y,
Li N, Qi J, Wang L, Shi Y, et al: Sp1 is involved in regulation of
cystathionine γ-lyase gene expression and biological function by
PI3K/Akt pathway in human hepatocellular carcinoma cell lines. Cell
Signal. 24:1229–1240. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Pathi S, Jutooru I, Chadalapaka G, Nair V,
Lee SO and Safe S: Aspirin inhibits colon cancer cell and tumor
growth and downregulates specificity protein (Sp) transcription
factors. PLoS One. 7:e482082012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Black AR, Black JD and Azizkhan-Clifford
J: Sp1 and kruppel-like factor family of transcription factors in
cell growth regulation and cancer. J Cell Physiol. 188:143–160.
2001. View
Article : Google Scholar : PubMed/NCBI
|
|
33
|
Li L, He S, Sun JM and Davie JR: Gene
regulation by Sp1 and Sp3. Biochem Cell Biol. 82:460–471. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Mukhopadhyay D and Datta K: Multiple
regulatory pathways of vascular permeability factor/vascular
endothelial growth factor (VPF/VEGF) expression in tumors. Semin
Cancer Biol. 14:123–130. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Liu S, Liu Z, Xie Z, Pang J, Yu J, Lehmann
E, Huynh L, Vukosavljevic T, Takeki M, Klisovic RB, et al:
Bortezomib induces DNA hypomethylation and silenced gene
transcription by interfering with Sp1/NF-kappaB-dependent DNA
methyltransferase activity in acute myeloid leukemia. Blood.
111:2364–2373. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Chadalapaka G, Jutooru I, Chintharlapalli
S, Papineni S, Smith R III, Li X and Safe S: Curcumin decreases
specificity protein expression in bladder cancer cells. Cancer Res.
68:5345–5354. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Pathi SS, Jutooru I, Chadalapaka G,
Sreevalsan S, Anand S, Thatcher GR and Safe S: GT-094, a NO-NSAID,
inhibits colon cancer cell growth by activation of a reactive
oxygen species-microRNA-27a: ZBTB10-specificity protein pathway.
Mol Cancer Res. 9:195–202. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Hsu TI, Wang MC, Chen SY, Huang ST, Yeh
YM, Su WC, Chang WC and Hung JJ: Betulinic acid decreases
specificity protein 1 (Sp1) level via increasing the sumoylation of
sp1 to inhibit lung cancer growth. Mol Pharmacol. 82:1115–1128.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Jia L, Huang Y, Zheng Y, Lyu M, Zhang C,
Meng Z, Gan Y and Yu G: miR-375 inhibits cell growth and correlates
with clinical outcomes in tongue squamous cell carcinoma. Oncol
Rep. 33:2061–2071. 2015.PubMed/NCBI
|
|
40
|
Jia LF, Huang YP, Zheng YF, Lyu MY, Wei
SB, Meng Z and Gan YH: miR-29b suppresses proliferation, migration,
and invasion of tongue squamous cell carcinoma through PTEN-AKT
signaling pathway by targeting Sp1. Oral Oncol. 50:1062–1071. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Qiu T, Zhou X, Wang J, Du Y, Xu J, Huang
Z, Zhu W, Shu Y and Liu P: MiR-145, miR-133a and miR-133b inhibit
proliferation, migration, invasion and cell cycle progression via
targeting transcription factor Sp1 in gastric cancer. FEBS Lett.
588:1168–1177. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Guo MM, Hu LH, Wang YQ, Chen P, Huang JG,
Lu N, He JH and Liao CG: miR-22 is down-regulated in gastric
cancer, and its overexpression inhibits cell migration and invasion
via targeting transcription factor Sp1. Med Oncol. 30:5422013.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Xu Y, Zhao F, Wang Z, Song Y, Luo Y, Zhang
X, Jiang L, Sun Z, Miao Z and Xu H: MicroRNA-335 acts as a
metastasis suppressor in gastric cancer by targeting Bcl-w and
specificity protein 1. Oncogene. 31:1398–1407. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Zhu X, Li Y, Xie C, Yin X, Liu Y, Cao Y,
Fang Y, Lin X, Xu Y, Xu W, et al: miR-145 sensitizes ovarian cancer
cells to paclitaxel by targeting Sp1 and Cdk6. Int J Cancer.
135:1286–1296. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Wang Y, Li M, Zang W, Ma Y, Wang N, Li P,
Wang T and Zhao G: MiR-429 up-regulation induces apoptosis and
suppresses invasion by targeting Bcl-2 and SP-1 in esophageal
carcinoma. Cell Oncol (Dordr). 36:385–394. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Zhang R, Luo H, Wang S, Chen W, Chen Z,
Wang HW, Chen Y, Yang J, Zhang X, Wu W, et al: MicroRNA-377
inhibited proliferation and invasion of human glioblastoma cells by
directly targeting specificity protein 1. Neuro Oncol.
16:1510–1522. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Cui W, Huang Z, He H, Gu N, Qin G, Lv J,
Zheng T, Sugimoto K and Wu Q: MiR-1188 at the imprinted Dlk1-Dio3
domain acts as a tumor suppressor in hepatoma cells. Mol Biol Cell.
26:1416–1427. 2015. View Article : Google Scholar : PubMed/NCBI
|