Introduction
Cutaneous squamous cell carcinoma (cSCC) is one of
the most common types of skin cancer leading to ~20% of annual skin
cancer-associated mortalities (1,2). Although
the risk of local recurrence and metastasis of cSCC are well
characterized, the molecular pathogenesis of this particular tumor
type remain unclear. As increasing numbers of mortalities occur due
to cSCC, it is urgent to clarify the molecular mechanisms of this
type of cancer and to develop novel and more effective treatment
strategies against this malignancy.
MicroRNA (miRNA), a class of naturally occurring,
17–25 nucleotide small noncoding RNA, regulates the expression of
genes through binding to the 3′ untranslated regions (3′-UTR) of
target mRNAs. MiRNAs have emerged as key factors involved in a
number of biological processes, including development,
differentiation, cell proliferation, and tumorigenesis (3–5). Previous
studies have shown that alterations in miRNA genes lead to tumor
formation, and miRNAs that regulate either tumor suppression or
tumor formation have been identified (6–8). Previous
studies have also identified a number of dysregulated miRNAs were
observed in cSCC (9,10). Zhou et al (11) demonstrated that miR-365 was
overexpressed in both cells and clinical specimens of cSCC
(11). The reduced expression of the
miR-193b/365a cluster observed during tumor progression suggests a
tumor suppressor role in cSCC (12).
MiR-199a inhibits cSCC cell proliferation and migration by
regulating CD44-Ezrin signaling (13).
Accumulating studies have shown that miR-31
expression is correlated with metastasis; however, the functional
role of this miRNA is extremely complex as it may function as an
oncogenic or a tumor-suppressive miRNA depending on the cellular
contexts (14–16). Previous studies have reported that
miR-31 is upregulated in cervical cancer (15,17,18), and
oesophageal squamous cell carcinoma (19), but downregulated in breast cancer
(20,21), bladder cancer (16), malignant mesothelioma (22), gastric cancer (23) and pancreatic cancer (24). Another study has demonstrated that
miR-31 is overexpressed in cSCC and that it regulates
cancer-associated phenotypes of cSCC (25), but the mechanisms behind its potential
involvement on proliferation and tumor cell invasion remain
unclear.
In the present study, the expression of miR-31 was
investigated in cSCC, and the downstream targets of miR-31 were
also explored. The role of miR-31 in cSCC was also analyzed in
relation to tumorigenesis and invasiveness.
Materials and methods
Cell culture and transfection
A cSCC cell line (A-431) and a normal skin cell line
(HaCaT) were obtained from the American type culture collection
(ATCC, Manassas, VA, USA) and cultivated in RPMI-1640 medium with
10% fetal bovine serum (both Gibco; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA). All cells were cultured in 95% air and 5%
CO2 at 37°C.
A-431 cells were seeded and transfected at a density
of 5×105 cells with miR-31 mimics or inhibitors (Qiagen
Operon, Alameda, CA, USA), RhoBTB1 siRNA and control siRNA using
Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.)
according to the manufacturer's instructions. A total of 24 or 48 h
later, the cells were collected and subjected to further
analysis.
RNA extraction and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted from transfected A-431 cells
using TRIzol reagent (Invitrogen, ThermoFisher Scientific, Inc.)
and then reverse-transcribed into cDNA. RT-qPCR was performed using
the SYBR Green qPCR Master Mix (Tiangen Biotech Co., Ltd., Beijing,
China) on an ABI 7300 PCR machine (Applied Biosystems, Inc.,
Foster, CA, USA). The sequences of the primers used to detect
miR-31 and U6 were as follows: miR-31, forward
5′-GGAGAGGCAAGATGCTGGCA-3′; U6, forward
5′-CGCAAGGATGACACGCAAATTC-3′; and a universal downstream reverse
primer, 5′-GTGCAGGGTCCGAGGT-3′. The primers used for detection of
RhoBTB1 were as follows: forward 5′-GGAGTGAAGGAGCCTGTGAG-3′; and
reverse 5′-TGCCAATGAACCCCTTACTC-3′. qPCR cycling conditions were as
follows: 95°C for 10 min, and then 95°C for 15 sec and 50°C for 2
min, for 40 cycles, followed by 60°C for 1 min. The melting curve
was 65–95°C. The relative mRNA expression levels were calculated as
2−∆∆Cq and were normalized against U6.
Luciferase reporter assays
A-431 cells were seeded into a 24-well plate at a
density of 2.5–3×104 cells/well), after 24 h the cells
were co-transfected with Renilla luciferase and luciferase reporter
plasmids containing miR-31 or vector control and the wild-type or
mutated target gene 3′-UTR using Lipofectamine 2000 (Invitrogen,
ThermoFisher Scientific, Inc.). A total of 48 h after transfection,
the luciferase activities were measured using a dual-luciferase
reporter assay system (Promega Corporation, Madison, WI, USA).
Firefly luciferase activities were normalized to Renilla luciferase
activity.
Western blotting
The cells were washed with phosphate-buffered saline
(PBS), and lysed with ice-cold RIPA (Sigma-Aldrich, St. Louis, MO,
USA). Total protein (60 μg) was extracted from transfected A-431
cells and separated on 10% SDS-polyacrylamide gels for RhoBTB1 and
β-actin detection. Anti-RhoBTB1 (catalog no. AV41883; 1:1,000
dilution) and anti-β-actin (catalog no. SAB2100037; 1:1,000
dilution) antibodies were purchased from Sigma-Aldrich. β-actin was
used as loading control. The protein in the gels was transferred to
nitrocellulose membranes, blocking was performed using 5% milk, and
then the membranes were incubated with the indicated antibodies in
recommended dilution overnight at 4°C. Then the membranes were
washed with 0.1 M PBST and incubated with HRP-conjugated secondary
antibody (goat anti-rabbit IgG, (H+L) HRP conjugate; catalog no.
A0545; Sigma-Aldrich). The signals were visualized using ECL
Substrates (GE Healthcare Life Sciences, Chalfont, UK), and
quantified using Optiquant software (Packard Instrument
Corporation, Meriden, CT, USA).
Cell viability assay
A cell viability assay was performed to investigate
the effect of miR-31 or RhoBTB1 expression on the proliferation of
A-431 cells. Following transfection as above, 6,000 cells of each
treatment group were plated in 96-well plates in triplicate, and
cell proliferation was assayed every 24 h using MTT (Beyotime
Institute of Biotechnology, Haimen, China) according to the
manufacturer's instructions.
Invasion assay
A-431 cells were cultivated to 80% confluence in
12-well plates. Then, we observed the procedures of cellular growth
at 24 h. Cells were seeded in the Transwell migration chamber
(Corning Inc., Corning, NY, USA) at a density of 2×106
cells and used to evaluate cell invasion. Then the cells that
invaded across the Matrigel-coated membrane were counted under a
light microscope (Olympus, Tokyo, Japan). All the experiments were
repeated in triplicate.
Statistical analysis
Data are expressed as the mean ± standard deviation
and analyzed by Student's t-test. Statistical analysis was
performed using SPSS 18.0 software (SPSS, Inc., Chicago, IL, USA).
P<0.05 was considered to indicate a statistically significant
difference.
Results
miR-31 expression is upregulated in
cSCC
A previous study revealed that miR-31 was
dysregulated in cSCC tissues (25),
therefore the present study examined miR-31 expression level in the
cSCC cell line A-431 by using RT-qPCR. As shown in Fig. 1, RT-qPCR results demonstrated that
compared with the HaCaT cell, miR-31 was significantly increased in
A-431 cells (P<0.01), which was in accordance with the previous
study (25). These results indicate
that miR-31 may be involved in cSCC tumor progression.
miR-31 affects human cSCC cell
viability and invasion
To further reveal the role of miR-31 in cSCC
development, miR-31 mimics or inhibitors were transfected into
A-431 cells to overexpress or silence miR-31 expression. As
demonstrated in Fig. 2A, following
transfection with miR-31 mimics, miR-31 expression was effectively
upregulated (P<0.01), and miR-31 expression was downregulated in
A-431 cells after tranfection with miR31 inhibitors (P<0.01). An
MTT assay demonstrated that overexpression of miR-31 significantly
increased cell viability and inhibition of miR-31 reduced viability
of A-431 cells (Fig. 2B), which
indicated that miR-31 contributed to cSCC tumorigenesis.
To verify the involvement of miR-31 in cSCC
tumorigenesis, a Transwell assay was performed to identify the
effect of miR-31 on cSCC cell invasion. The results demonstrated
that the invasion capabilities of A-431 cells was markedly
increased in the miR-31 mimics group (P<0.01) and reduced in the
miR-31 inhibitor group (P<0.01), indicating that miR-31 may
induce A-431 cell invasion (Fig. 3C).
In conclusion, the results demonstrated that miR-31 contributed to
cSCC cell viability and invasion, which further indicated that
miR-31 may be involved cSCC development.
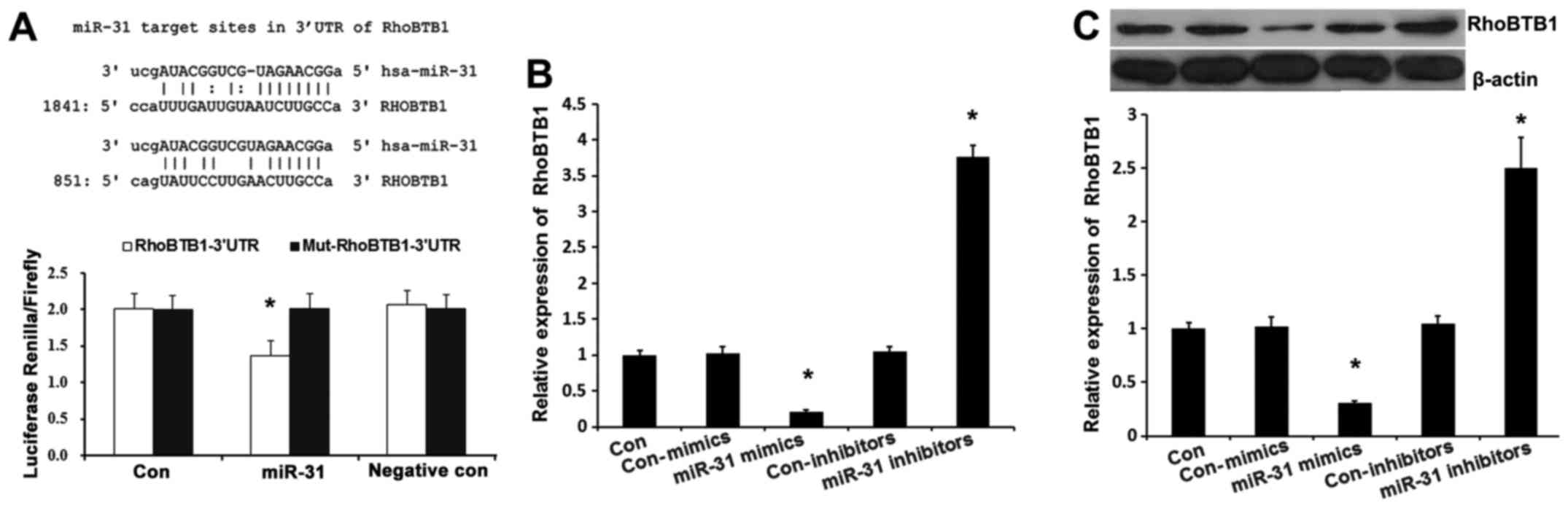
RhoBTB1 is a direct target of miR-31
in cSCC
In order to elucidate the underlying molecular
mechanism of miR-31 action, a bioinformatic analysis was performed
using mirco-RNA.org (http://www.microrna.org/microrna/home.do) to predict
the possible target genes of miR-31. It was demonstrated that
RhoBTB1 contained two theoretical miR-31 binding sites in its 3′
UTR (Fig. 3A). To demonstrate whether
RhoBTB1 was directly targeted by miR-31, a luciferase reporter gene
assay was performed in A-431 cells. As presented in Fig. 3B, co-transfection of miR-31 suppressed
the luciferase activity of the reporter containing the wild-type
RhoBTB1 3′-UTR sequence, but failed to inhibit that of mutated
RhoBTB1 by dual-luciferase reporter assay. These data indicated
that miR-31 could directly target the 3′-UTR sequences of RhoBTB1.
Additionally, in A-431 cells, the expression of RhoBTB1 was
suppressed by miR-31 mimics transfection (Fig. 3B; P<0.01), while RhoBTB1 expression
was enhanced by miR-133a inhibitor, which was also confirmed by
western blot analysis (Fig. 3C;
P<0.01). These results demonstrated that endogenous RhoBTB1
expression is directly targeted and regulated by miR-31 and
suggested that miR-31 upregulation in cSCC may reduce the
expression of RhoBTB1.
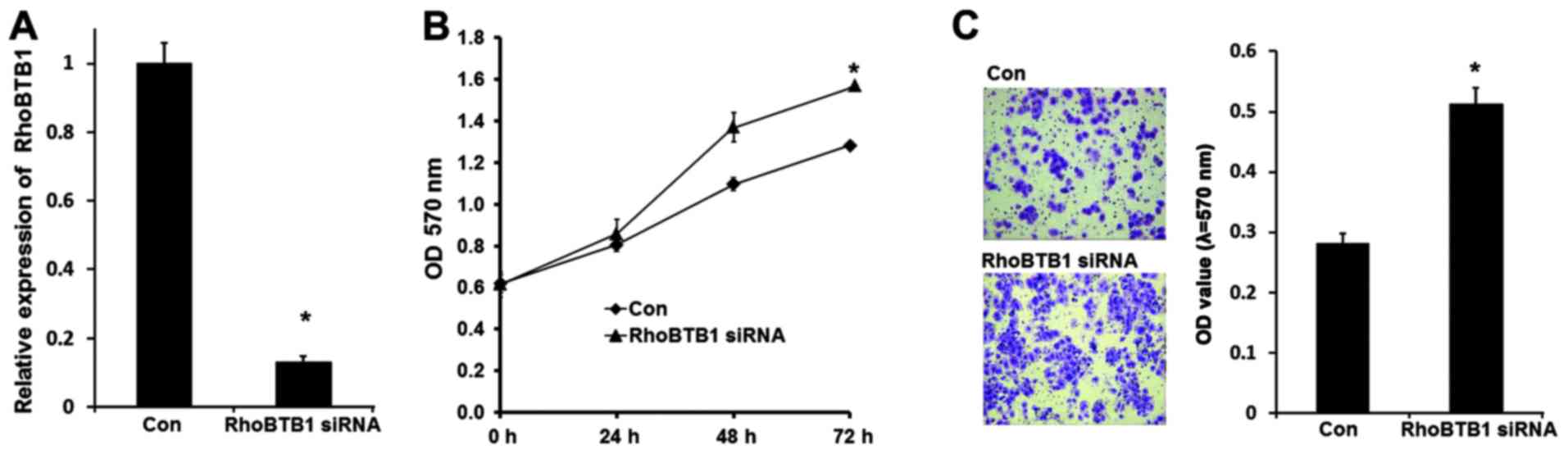
Inhibition of RhoBTB1 is responsible
for the tumor promoting effects of miR-31
To further confirm that miR-31-induced cSCC
progression is mediated by RhoBTB1, RhoBTB1 expression was knocked
down in A-431 cells by siRNA. As shown in Fig. 4A, RhoBTB1 mRNA was effectively
inhibited after RhoBTB1 siRNA were transfected, and the MTT assay
results demonstrated that A-431 cell proliferation was induced with
suppression of RhoBTB1. Transwell invasion assay results
demonstrated that inhibition of RhoBTB1 promoted A-431 cell
invasion (Fig. 4B). These data
indicated that miR-31 promoted tumor development at least partly
through suppressing tumor supressor RhoBTB1.
Discussion
Over the last decade, accumulating evidence has
demonstrated that miRNAs are involved in the pathogenesis of a
number of human diseases, including cancer. miR-31 may act as an
oncogenic or a tumour-suppressive miRNA and serves important roles
in tumorigenesis and the progression of chemotherapy resistance
(14,22,26). For
example, downregulation of miR-31 confers resistance to
chemotherapy-induced apoptosis in prostate cancer cells (26), and it has been reported that miR-31
acts as an oncogenic miRNA (oncomir) in lung cancer by targeting
specific tumor suppressors LATS2 and PPP2R2A (14). MiRNA-31 functions as an oncogenic
MicroRNA in human colorectal cancer by repressing RAS p21 GTPase
activating protein 1 (RASA1), SATB2 and HIF-1α (FIH-1) (15,18,27). In
the present study, it was demonstrated that miR-31 was
significantly upregulated in cSCC. This result was consistent with
the findings of previous studies that miR-31 is overexpression in
cSCC and induced cancer-associated phenotypes of cSCC (25). To examine the effect of miR-31 on cSCC
proliferation and invasion, miR-31 mimics and inhibitors were
transfected into A-431 cells to overexpress and knockdown miR-31.
MTT assay results showed that A-431 cell proliferation was
increased after miR-31 mimics transfection and decreased after
miR-31 inhibitor transfection (Fig.
2A). The ability of cell invasion was greatly increased by
miR-31 mimics and decreased by miR-31 inhibitor (Fig. 2B). These results suggest that miR-31
acts primarily as an oncogene in cSCC.
RhoTBT1 belongs to RhoBTB subfamily which are
atypical members of the Rho family of small GTPases. The RhoBTB
subfamily is composed of three members, RhoTBT1, RhoTBT2 and
RhoTBT3 (28). RhoTBT2 may act as a
tumor suppressor; it has been reported that lack of RHOBTB2
transcripts results in growth inhibition in breast cancer (28). Previous studies have also found high
rates of loss of heterozygosity at the RHOTBT2 locus in gastric
tumors and bladder tumors (29,30).
Similarly to RhoTBT2, RhoTBT1 was also recently reported to be a
tumor supressor in a study on head and neck cancer and colon cancer
(31,32). However, analysis of RhoTBT2 in cSCC
has not yet been reported. In the present study, RhoTBT2 was also
identified to be a direct target of miR-31 in cSCC and miR-31
upregulation in cSCC might suppress RhoBTB1 expression. To further
examine whether the depressed RhoBTB1 was responsible for the tumor
promoting effects of miR-31, RhoBTB1 was silenced by siRNA, as
indicated in Fig. 4, suppression of
RhoTBT1 in A-431 induced cell proliferation, which was consistent
with the function of miR-31 mimics. The knockdown of RhoTBT1 also
promoted A-431 cells invasion.
In conclusion, the present study suggests high
levels of miR-31 are involved in cSCC tumorigenesis, and tumor
supressor RhoBTB1 was identified as a direct target of miR-31.
Overexpression of miR-31 promotes tumor proliferation through
reducing the expression of RhoBTB1. These observations shed new
light on mechanisms underlying development of cSCC and supply
potential novel therapeutic targets in inhibiting cSCC
tumorigenesis.
References
|
1
|
Alam M and Ratner D: Cutaneous
squamous-cell carcinoma. N Engl J Med. 344:975–983. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ratushny V, Gober MD, Hick R, Ridky TW and
Seykora JT: From keratinocyte to cancer: The pathogenesis and
modeling of cutaneous squamous cell carcinoma. J Clin Invest.
122:464–472. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bushati N and Cohen SM: MicroRNA
functions. Annu Rev Cell Dev Biol. 23:175–205. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zhang Y, Yang Q and Wang S: MicroRNAs: A
new key in lung cancer. Cancer Chemother Pharmacol. 74:1105–1111.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Xue J, Niu J, Wu J and Wu ZH: MicroRNAs in
cancer therapeutic response: Friend and foe. World J Clin Oncol.
5:730–743. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kent OA and Mendell JT: A small piece in
the cancer puzzle: MicroRNAs as tumor suppressors and oncogenes.
Oncogene. 25:6188–6196. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Acunzo M, Romano G, Wernicke D and Croce
CM: MicroRNA and cancer-a brief overview. Adv Biol Regul. 57:1–9.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lee JW, Choi CH, Choi JJ, Park YA, Kim SJ,
Hwang SY, Kim WY, Kim TJ, Lee JH, Kim BG and Bae DS: Altered
MicroRNA expression in cervical carcinomas. Clin Cancer Res.
14:2535–2542. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Bruegger C, Kempf W, Spoerri I, Arnold AW,
Itin PH and Burger B: MicroRNA expression differs in cutaneous
squamous cell carcinomas and healthy skin of immunocompetent
individuals. Exp Dermatol. 22:426–428. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Sand M, Skrygan M, Georgas D, Sand D, Hahn
SA, Gambichler T, Altmeyer P and Bechara FG: Microarray analysis of
microRNA expression in cutaneous squamous cell carcinoma. J
Dermatol Sci. 68:119–126. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhou M, Liu W, Ma S, Cao H, Peng X, Guo L,
Zhou X, Zheng L, Guo L, Wan M, et al: A novel onco-miR-365 induces
cutaneous squamous cell carcinoma. Carcinogenesis. 34:1653–1659.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Gastaldi C, Bertero T, Xu N,
Bourget-Ponzio I, Lebrigand K, Fourre S, Popa A, Cardot-Leccia N,
Meneguzzi G, Sonkoly E, et al: MiR-193b/365a cluster controls
progression of epidermal squamous cell carcinoma. Carcinogenesis.
35:1110–1120. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wang SH, Zhou JD, He QY, Yin ZQ, Cao K and
Luo CQ: MiR-199a inhibits the ability of proliferation and
migration by regulating CD44-Ezrin signaling in cutaneous squamous
cell carcinoma cells. Int J Clin Exp Pathol. 7:7131–7141.
2014.PubMed/NCBI
|
|
14
|
Liu X, Sempere LF, Ouyang H, Memoli VA,
Andrew AS, Luo Y, Demidenko E, Korc M, Shi W, Preis M, et al:
MicroRNA-31 functions as an oncogenic microRNA in mouse and human
lung cancer cells by repressing specific tumor suppressors. J Clin
Invest. 120:1298–1309. 2010. View
Article : Google Scholar : PubMed/NCBI
|
|
15
|
Sun D, Yu F, Ma Y, Zhao R, Chen X, Zhu J,
Zhang CY, Chen J and Zhang J: MicroRNA-31 activates the RAS pathway
and functions as an oncogenic MicroRNA in human colorectal cancer
by repressing RAS p21 GTPase activating protein 1 (RASA1). J Biol
Chem. 288:9508–9518. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wang S, Li Q, Wang K, Dai Y, Yang J, Xue
S, Han F, Zhang Q, Liu J and Wu W: Decreased expression of
microRNA-31 associates with aggressive tumor progression and poor
prognosis in patients with bladder cancer. Clin Transl Oncol.
15:849–854. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Xu XM, Qian JC, Deng ZL, Cai Z, Tang T,
Wang P, Zhang KH and Cai JP: Expression of miR-21, miR-31, miR-96
and miR-135b is correlated with the clinical parameters of
colorectal cancer. Oncol Lett. 4:339–345. 2012.PubMed/NCBI
|
|
18
|
Yang MH, Yu J, Chen N, Wang XY, Liu XY,
Wang S and Ding YQ: Elevated microRNA-31 expression regulates
colorectal cancer progression by repressing its target gene SATB2.
PLoS One. 8:e853532013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhang T, Wang Q, Zhao D, Cui Y, Cao B, Guo
L and Lu SH: The oncogenetic role of microRNA-31 as a potential
biomarker in oesophageal squamous cell carcinoma. Clin Sci (Lond).
121:437–447. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Viré E, Curtis C, Davalos V, Git A, Robson
S, Villanueva A, Vidal A, Barbieri I, Aparicio S, Esteller M, et
al: The breast cancer oncogene EMSY represses transcription of
antimetastatic microRNA miR-31. Mol Cell. 53:806–818. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Körner C, Keklikoglou I, Bender C, Wörner
A, Münstermann E and Wiemann S: MicroRNA-31 sensitizes human breast
cells to apoptosis by direct targeting of protein kinase C epsilon
(PKCepsilon). J Biol Chem. 288:8750–8761. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ivanov SV, Goparaju CM, Lopez P, Zavadil
J, Toren-Haritan G, Rosenwald S, Hoshen M, Chajut A, Cohen D and
Pass HI: Pro-tumorigenic effects of miR-31 loss in mesothelioma. J
Biol Chem. 285:22809–22817. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Guo J, Miao Y, Xiao B, Huan R, Jiang Z,
Meng D and Wang Y: Differential expression of microRNA species in
human gastric cancer versus non-tumorous tissues. J Gastroenterol
Hepatol. 24:652–657. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Papaconstantinou IG, Manta A, Gazouli M,
Lyberopoulou A, Lykoudis PM, Polymeneas G and Voros D: Expression
of microRNAs in patients with pancreatic cancer and its prognostic
significance. Pancreas. 42:67–71. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wang A, Landén NX, Meisgen F,
Lohcharoenkal W, Ståhle M, Sonkoly E and Pivarcsi A: MicroRNA-31 is
overexpressed in cutaneous squamous cell carcinoma and regulates
cell motility and colony formation ability of tumor cells. PLoS
One. 9:e1032062014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Bhatnagar N, Li X, Padi SK, Zhang Q, Tang
MS and Guo B: Downregulation of miR-205 and miR-31 confers
resistance to chemotherapy-induced apoptosis in prostate cancer
cells. Cell Death Dis. 1:e1052010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Chen T, Yao LQ, Shi Q, Ren Z, Ye LC, Xu
JM, Zhou PH and Zhong YS: MicroRNA-31 contributes to colorectal
cancer development by targeting factor inhibiting HIF-1α (FIH-1).
Cancer Biol Ther. 15:516–523. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Hamaguchi M, Meth JL, von Klitzing C, Wei
W, Esposito D, Rodgers L, Walsh T, Welcsh P, King MC and Wigler MH:
DBC2, a candidate for a tumor suppressor gene involved in breast
cancer. Proc Natl Acad Sci USA. 99:13647–13652. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Knowles MA, Aveyard JS, Taylor CF, Harnden
P and Bass S: Mutation analysis of the 8p candidate tumour
suppressor genes DBC2 (RHOBTB2) and LZTS1 in bladder cancer. Cancer
Lett. 225:121–130. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Cho YG, Choi BJ, Kim CJ, Song JH, Zhang C,
Nam SW, Lee JY and Park WS: Genetic analysis of the DBC2 gene in
gastric cancer. Acta Oncol. 47:366–371. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Beder LB, Gunduz M, Ouchida M, Gunduz E,
Sakai A, Fukushima K, Nagatsuka H, Ito S, Honjo N, Nishizaki K and
Shimizu K: Identification of a candidate tumor suppressor gene
RHOBTB1 located at a novel allelic loss region 10q21 in head and
neck cancer. J Cancer Res Clin Oncol. 132:19–27. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Xu RS, Wu XD, Zhang SQ, Li CF, Yang L, Li
DD, Zhang BG, Zhang Y, Jin JP and Zhang B: The tumor suppressor
gene RhoBTB1 is a novel target of miR-31 in human colon cancer. Int
J Oncol. 42:676–682. 2013.PubMed/NCBI
|