Introduction
Colorectal cancer (CRC) is the fourth most common
cancer and the second leading cause of cancer-associated mortality
in the United States. Although the incidence and mortality rates of
CRC have decreased in recent decades as a result of earlier
diagnosis, >10,000 new cases are diagnosed annually in the USA
(1).
The efficacy of CRC treatment is dependent on the
preoperative condition of patients, which includes the local depth
of tumor incision (T stage), lymph-node metastasis (N stage) and
the histological grade of tumors (1).
Although useful, in certain cases these factors fail to
differentiate tumors from each other, which is important for CRC
patients with lymph node metastasis that require adjuvant
chemotherapy following curative resection (1). However, certain patients may not benefit
from additional therapy (1). As a
result, the identification of a biomarker that accurately
corresponds with the various stages of CRC is urgently
required.
Dysfunction of the cellular transport machinery is
common in cancer cells, and this observation has led to the
development of therapies that target this machinery (2). Karyopherin α (KPNA) belongs to a family
of nuclear transport proteins, which interact with cellular cargo
via the nuclear localization signal (3,4). In
addition to combined action, importin α also binds the nuclear
cargoes directly without the assistance of importin β (5). As a member of the KPNA family, KPNA2 is
a 58 kDa protein composed of 529 amino acids (5,6). A change
of bowel habit and blood in the stool are most common symptoms in
patients with CRC. Although the cause of CRC is remains unclear,
certain health history can affect the risk of developing CRC,
including a family history of CRC, certain hereditary conditions
(Lynch syndrome), a history of ulcerative colitis and polyps in the
colon and rectum.
A number of studies have demonstrated that KPNA2 is
expressed at a high level in a variety of types of cancer,
including breast and lung (7–11). However, few studies have investigated
the expression of KPNA2 in CRC. Therefore, the aim of the present
study was to analyze KPNA2 expression in malignant colorectal
tumors and to elucidate its function as an oncogene in CRC.
Materials and methods
Ethics statement
All protocols were reviewed and approved by the
Ethical Committee of Harbin Medical University (Harbin, China) and
written informed consent was obtained from all participants.
Patients and tissue preparation
A total of 30 CRC patients (15 males and 15 females;
mean age, 56.4 years) diagnosed with colorectal adenocarcinoma
(stage I–IV), who received surgical treatment at the Second
Affiliated Hospital of Harbin Medical University (Harbin, China)
between January 2014 and June 2014, were enrolled in the present
study, as well as 30 healthy volunteers (15 males and 15 females;
mean age, 59.1 years). None of the patients had previously
undergone neoadjuvant treatment. For enzyme-linked immunosorbent
assay (ELISA), preoperative venous blood (10 ml) was collected from
each participant and centrifuged (1006.2 × g) at 4°C for
serum collection. The serum samples collected were stored at −80°C
until additional analysis occurred. For reverse
transcription-quantitative polymerase chain reaction (RT-qPCR),
cancerous and paired normal tissues (resected 6-cm from the
cancerous tissues) were collected from each participant. For RNA
extraction, the specimens resected during surgery were immediately
snap-frozen in liquid nitrogen and subsequently stored at −80°C,
followed by fixing in 10% buffered formalin (Sigma Aldrich; EMD
Millipore, Billerica, MA, USA) for 24 h and embedding in paraffin
(Sigma-Aldrich; EMD Millipore). To investigate the association
between KPNA2 expression, clinicopathological features and survival
of CRC patients following radical surgery, a larger sample size was
required. Therefore, an additional 300 patients with CRC that
underwent radical resection at The Second Affiliated Hospital of
Harbin Medical University (Harbin, China) between January 2007 and
December 2008 were included in the present study. Patients that had
received preoperative chemotherapy or irradiation were excluded.
Tumors were staged according to the Union for International Cancer
Control staging system. The patient cohort included 175 men and 125
women (mean age, 62.1 years; range, 23–83 years). Cancerous tissues
and paired normal tissues were excised and fixed in 10% buffered
formalin for 24 h and embedded in paraffin blocks. For all samples,
histological diagnosis was performed by three independent and
experienced pathologists.
Immunohistochemical analysis
Tissue samples underwent immunohistochemical
analysis using the avidin-biotin-peroxidase method. Sections were
deparaffinized in xylene (Sigma-Aldrich; EMD Millipore) and
dehydrated using graded alcohol prior to endogenous peroxidase
activity blocking using 0.5% hydrogen peroxide (Sigma-Aldrich; EMD
Millipore) in methanol (Sigma-Aldrich; EMD Millipore) for 10 min.
Non-specific binding was blocked by incubating tissue sections with
10% normal goat serum (Sigma-Aldrich; EMD Millipore) in
phosphate-buffered saline (PBS) for 1 h at room temperature. The
sections were subsequently incubated with polyclonal human KPNA2
antibody (dilution, 1:300; catalog no., 10819-1-AP; Proteintech,
Inc., Wuhan, China) in PBS at 4°C overnight, followed by 1 h of
incubation with biotinylated goat anti-mouse immunoglobulin G (IgG;
ZDR 5210; dilution, 1:400; ZSGB-BIO, Beijing, China) at room
temperature. Subsequently, sections were treated with
streptavidin-peroxidase for 10 min at room temperature
(Sigma-Aldrich; EMD Millipore). Sections were then incubated with
0.1% 3,3′-diaminobenzidine (ZSGB-BIO) in PBS with 0.05% hydrogen
peroxide for 5 min at room temperature. All tissue specimens were
assessed separately by two pathologists blinded to the
clinicopathological data. KPNA2 expression in CRC specimens was
evaluated microscopically (Nikon ECLIPSE Ti-E; Nikon Corporation,
Tokyo, Japan) at low magnification (×40) and the results were
confirmed at high magnifications (×200 and ×400) by two surgical
pathologists blinded to the clinical data. An immunoreactivity
scoring system was applied, according to previously reported
procedures (12). The percentage of
positive cells was scored as follows: 0 (<5%, negative); 1
(5–25%, sporadic); 2 (25–50%, focal); and 3 (>50%, diffuse). The
staining intensity was scored as follows: 0, no staining; 1, weak
staining; 2, moderate staining; and 3, strong staining. The KPNA2
immunostaining score was calculated using the following formula:
Immunostaining score = positive cell score × staining intensity
score. The immunostaining score ranged between 0 and 9. A score of
≥4 indicated high KPNA2 expression.
RT-qPCR
Total RNA was extracted from tissue samples using
TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA) according to the manufacturer's protocol.
First-strand complementary DNA (cDNA) was synthesized using 1 µg
total RNA and a RevertAid™H Minus First Strand cDNA Synthesis kit
(Fermentas Inc., Burlington, ON, Canada) according to the
manufacturer's protocol. The primers for KPNA2 and
glyceraldehyde-3-phosphate dehydrogenase (GAPDH; internal control)
were obtained from Takara Biotechnology Co., Ltd., (Dalian, China),
with the following sequences: forward 5′-CAAGGCTGTGGTAGATGG-3′ and
reverse, 5′-GCGGCAAAGATTAGAAAG-3′ for KPNA2; forward,
5′-CAATGACCCCTTCATTGACC-3′ and reverse, 5′-GACAAGCTTCCCGTTCTCAG-3′
for GAPDH. The KPNA2 and GAPDH genes were amplified from the cDNA
pool using gene-specific primers and the Power SYBR Green PCR
Master Mix (Applied Biosystems; Thermo Fisher Scientific, Inc.) in
an ABI PRISM 7500 Sequence Detection System (Applied Biosystems;
Thermo Fisher Scientific, Inc.). PCR was performed under the
following conditions: Initial denaturation at 95°C for 5 min
followed by 40 cycles of 94°C for 20 sec, 58°C for 20 sec and 72°C
for 20 sec. RT-qPCR was performed at least three times, and a
negative control group was also used. KPNA2 and GAPDH were
amplified in an identical reaction. The expression of the target
gene was evaluated using the 2−ΔΔCq method for relative
quantification using GAPDH as the internal reference gene (7).
Fluorometric sandwich ELISA
KPNA2 protein levels in human serum were determined
using the KPNA2 ELISA kit (Shanghai BlueGene Biotech Co., Ltd.,
Shanghai, China). The serum from patients and healthy volunteers
were incubated with a KPNA2-horseradish peroxidase (HRP) conjugate
(dilution, 1:5,000; ZSR 5210) in a pre-coated plate (coated with
HRP-conjugated rabbit antibody) at 37°C for 1 h. Following
incubation, the wells were emptied and washed 5 times with wash
solution (PBS and Tween 20). The wells were subsequently incubated
with a substrate for HRP enzyme at 37°C for 15 min and subsequently
a blue-colored complex was formed due to the enzyme-substrate
reaction. Finally, a stop solution (1M H2SO4)
was added to terminate the reaction, as signified by a color change
from blue to yellow. The intensity of the yellow color was measured
spectrophotometrically at a wavelength of 450 nm using a Thermo
Multiskan MK3 microplate reader (Thermo Fisher Scientific, Inc.).
The intensity of the color was inversely proportional to the KPNA2
concentration: As KPNA2 binds more sites in the serum, fewer sites
remain for KPNA2-HRP conjugate binding. A standard curve was
plotted correlating the intensity of the color (optical density)
with the concentration. The KPNA2 concentration in each sample was
interpolated based on the standard curve.
Statistical analysis
Data were analyzed using SPSS version 17.0 (SPSS
Inc., Chicago, IL, USA). All continuous variables were expressed as
the mean ± standard deviation. The nonparametric Mann-Whitney U
test was utilized to analyze variations in ELISA results.
χ2 test was applied to evaluate the statistical
significance of the association between KPNA2 expression and other
clinicopathological variables. For univariate survival analysis,
survival curves were calculated according to the Kaplan-Meier
method. The difference in survival rates was assessed using the
log-rank test. Cox's proportional hazard model was utilized to
identify factors that exhibited a significant influence on
survival. P<0.05 was considered to indicate a statistically
significant difference.
Results
KPNA2 is overexpressed in CRC tissues
compared with paired normal tissues
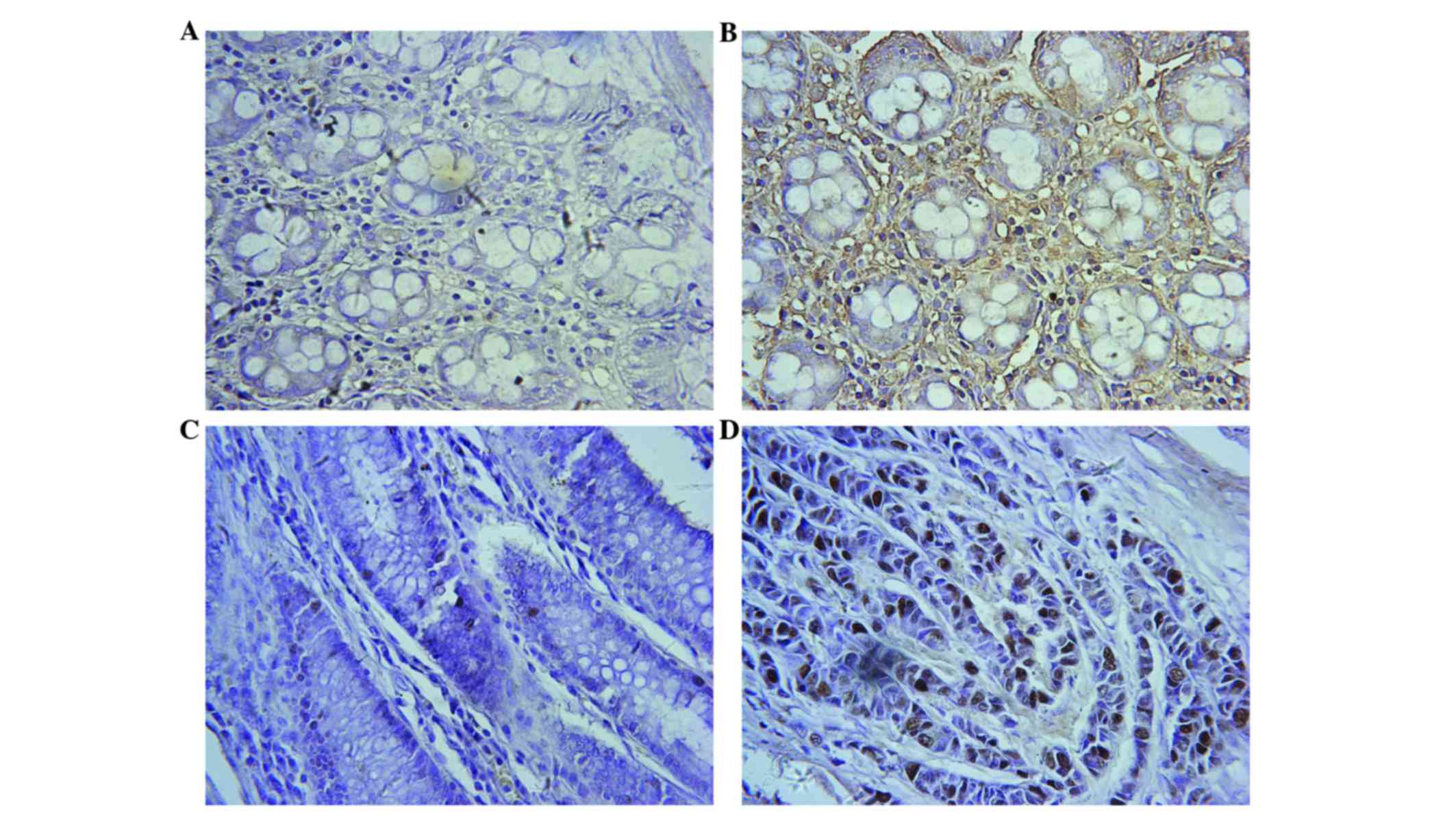
Immunohistochemistry identified nuclear KPNA2
expression in the majority of cells of the cancer tissues examined
(26/30 samples), and a small number of paired normal tissues
(Fig. 1). In general, CRC tissues
exhibited a markedly increased KPNA2 expression rate compared with
paired normal tissues. KPNA2 expression was identified in 272/300
(90.7%) CRC tissues, and low KPNA2 expression was observed in
44/300 (14.7%) paired normal tissues (P<0.001; Table I), while no KPNA2 expression was
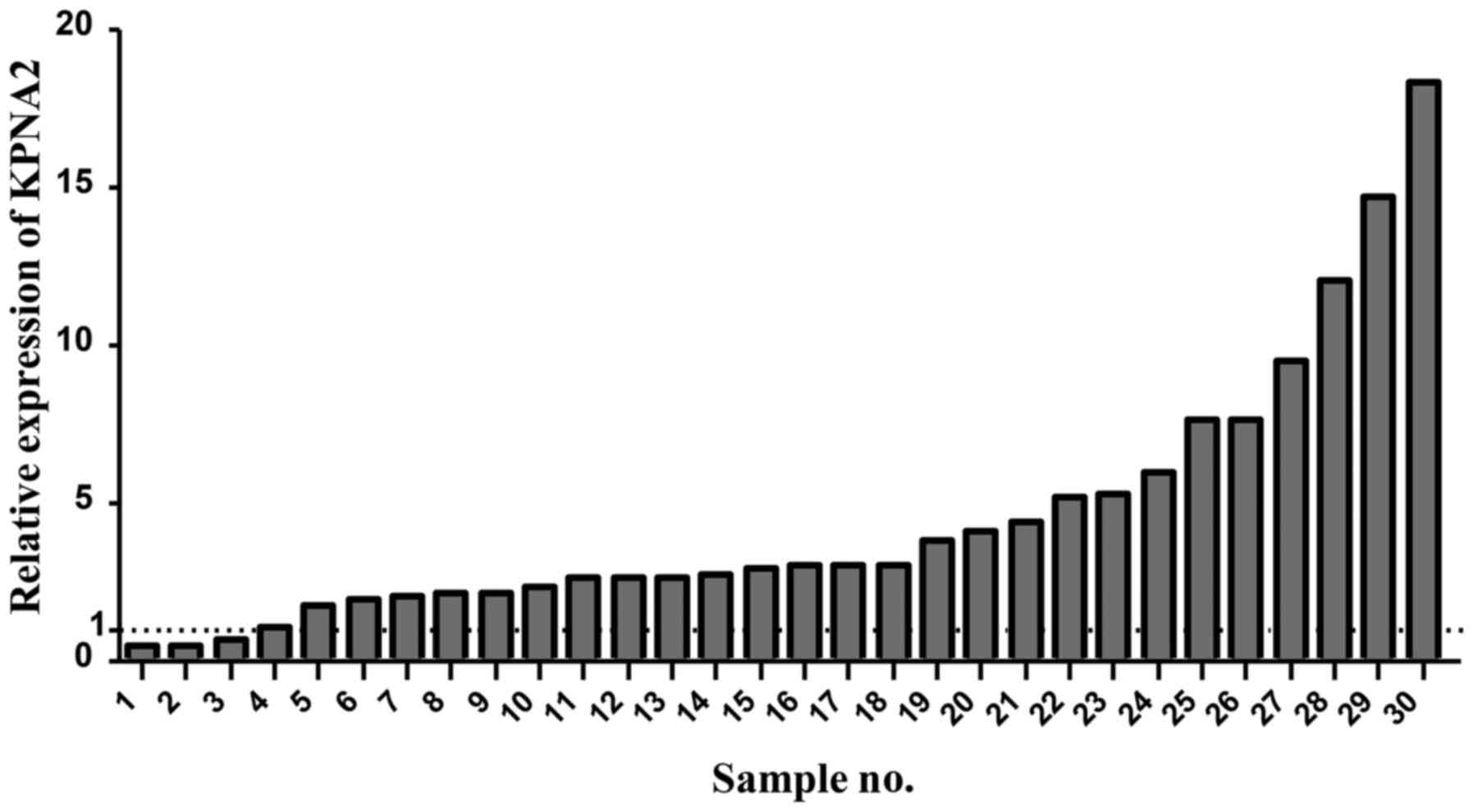
observed in 256/300 paired normal tissues (Fig. 1A). KPNA2 expression in 30 human CRC
tissues and paired normal tissues was measured by RT-qPCR. A total
of 26/30 (86.7%) CRC samples exhibited increased KPNA2 expression
compared with normal tissues. Relative KPNA2 expression was
significantly increased in colorectal adenocarcinoma tissues
compared with paired normal tissues (P<0.001; Fig. 2). These results demonstrate that KPNA2
expression is significantly increased in cancer tissues compared
with paired normal tissues.
 | Table I.KPNA2 expression in 300 colorectal
cancer and paired normal tissues. |
Table I.
KPNA2 expression in 300 colorectal
cancer and paired normal tissues.
|
| KPNA2
expressiona, n (%) |
|
|
|---|
|
|
|
|
|
|---|
| Tissue type | Positive | Negative | χ2 |
P-valueb |
|---|
| Colorectal
cancer | 272 (90.7) | 28 (9.3) | 347.55 | <0.001 |
| Paired normal | 44
(14.7) | 256 (85.3) |
|
|
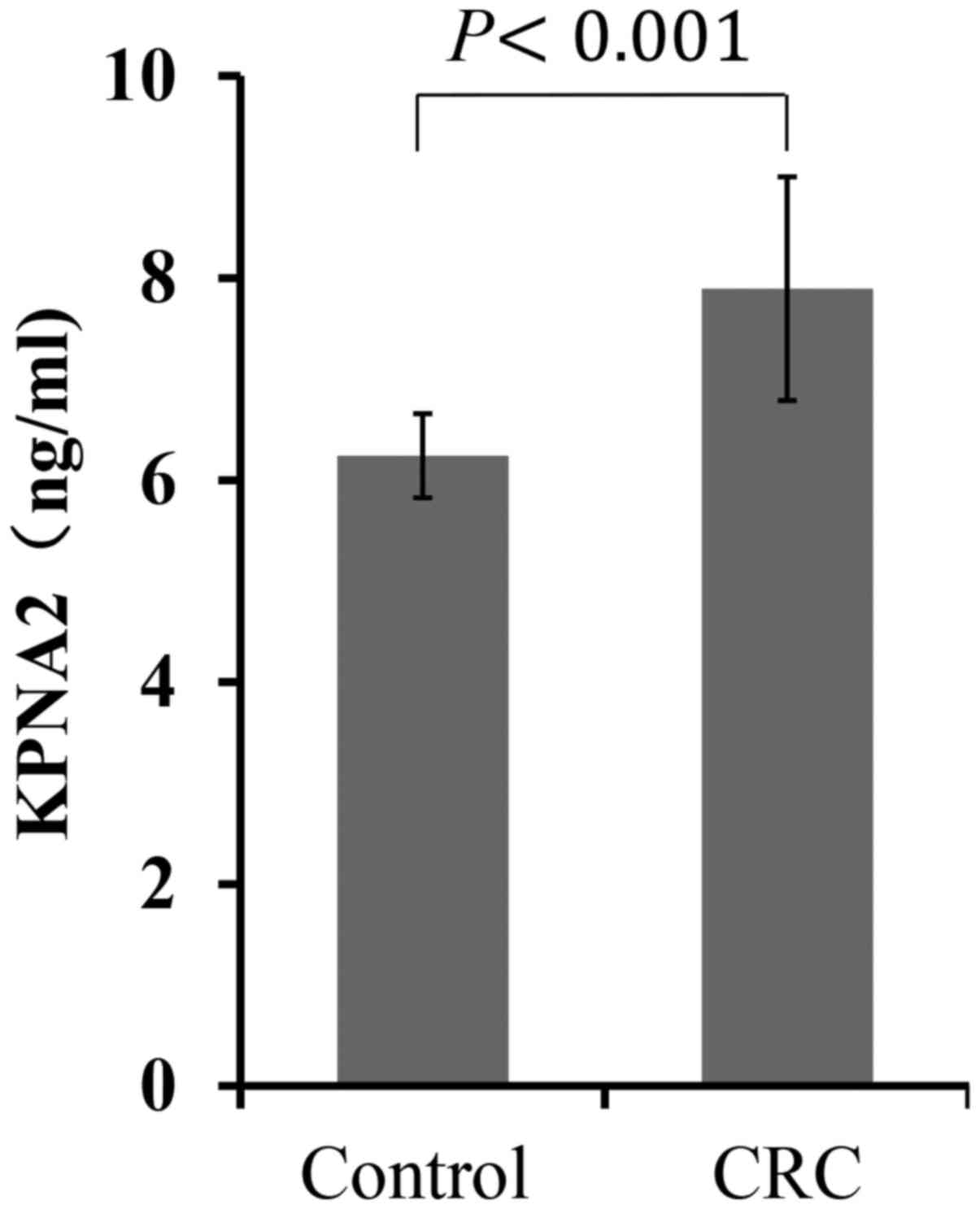
KPNA2 serum levels are increased in
CRC patients
To investigate the preoperative diagnostic value of
KPNA2 expression for CRC, ELISA was used to analyze KPNA2
expression in the serum of 30 CRC patients and 10 healthy
individuals. As shown in Fig. 3, the
serum KPNA2 levels were significantly increased in CRC patients
compared with healthy individuals (7.84±1.11 ng/ml vs. 6.24±0.42
ng/ml; P<0.001; Fig. 3).
Correlation between KPNA2 expression
and patient clinicopathological features
The associations between KPNA2 expression and
clinicopathological features, including age, gender, tumor size,
differentiation, Tumor-Node-Metastasis (TNM) stage, infiltration
depth, lymph node involvement, lymphovascular invasion (LVI),
perineural invasion (PNI) and tumor location of CRC patients, are
shown in Table II. High KPNA2
expression was significantly associated with lymph node involvement
(P<0.001), LVI and PNI (P<0.001) in CRC patients. In
addition, KPNA2 expression was significantly associated with tumor
differentiation (P=0.020), infiltration depth (P=0.012) and TNM
stage (P=0.001). The results additionally demonstrated that the
intensity of KPNA2 expression was increased in late-stage patients
compared with those with early-stage CRC. However, no significant
differences were identified between KPNA2 expression and age
(P=0.285), gender (P=0.358), tumor size (P=0.457) or tumor location
(P=0.312) in CRC patients.
 | Table II.Associations between KPNA2 expression
and patient clinicopathological features were analyzed in 300
colorectal cancer patients. |
Table II.
Associations between KPNA2 expression
and patient clinicopathological features were analyzed in 300
colorectal cancer patients.
| Parameter | High KPNA2
expressiona, n (%) | Low KPNA2
expressiona, n (%) | χ2 | P-value |
|---|
| Gender |
|
|
|
|
|
Male | 110 (36.67) | 65 (21.67) | 0.84 | 0.358b |
|
Female | 72 (24.00) | 53 (17.67) |
|
|
| Age, years |
|
|
|
|
|
≤60 | 78 (26.00)) | 58 (19.33) | 1.15 | 0.285b |
|
>60 | 104 (34.67) | 60 (20.00) |
|
|
| Tumor size, cm |
|
|
|
|
|
<5 | 117 (39.00) | 81 (27.00) | 0.61 | 0.457b |
| ≥5 | 65
(21.67) | 37 (12.33) |
|
|
|
Differentiation |
|
|
|
|
|
Well | 15 (5.00) | 26 (8.67) | 12.56 | 0.020b,d |
|
Moderate | 119 (39.67) | 71
(23.67) |
|
|
|
Poor | 48 (16.00) | 21 (7.00) |
|
|
| TNM stage |
|
|
|
|
| I | 17 (5.67) | 20 (6.67) | 16.86 | 0.001c,d |
| II | 85 (28.33) | 69 (23.00) |
|
|
|
III | 53 (17.67) | 25 (8.33) |
|
|
| IV | 27 (9.00) | 4 (1.33) |
|
|
| Infiltration
depth |
|
|
|
|
|
T1+T2 | 23 (7.67) | 29 (9.67) | 6.66 | 0.012b,d |
|
T3+T4 | 155 (51.67) | 89 (30.07) |
|
|
| Lymph node
involvement, n |
|
|
|
|
| 0 | 102 (34.00) | 97 (32.33) | 21.94 |
<0.001b,d |
| ≥1 | 80 (26.67) | 21 (7.00) |
|
|
| LVI or PNI |
|
|
|
|
|
Negative | 65 (21.67) | 67 (22.33) | 12.89 |
<0.001b,d |
|
Positive | 117 (39.00) | 51 (17.00) |
|
|
| Tumor location |
|
|
|
|
|
Right-side colon | 69 (23.00) | 42 (14.00) | 2.38 | 0.312b |
|
Left-side colon | 60 (20.00) | 32 (10.67) |
|
|
|
Rectum | 53 (17.67) | 44 (14.67) |
|
|
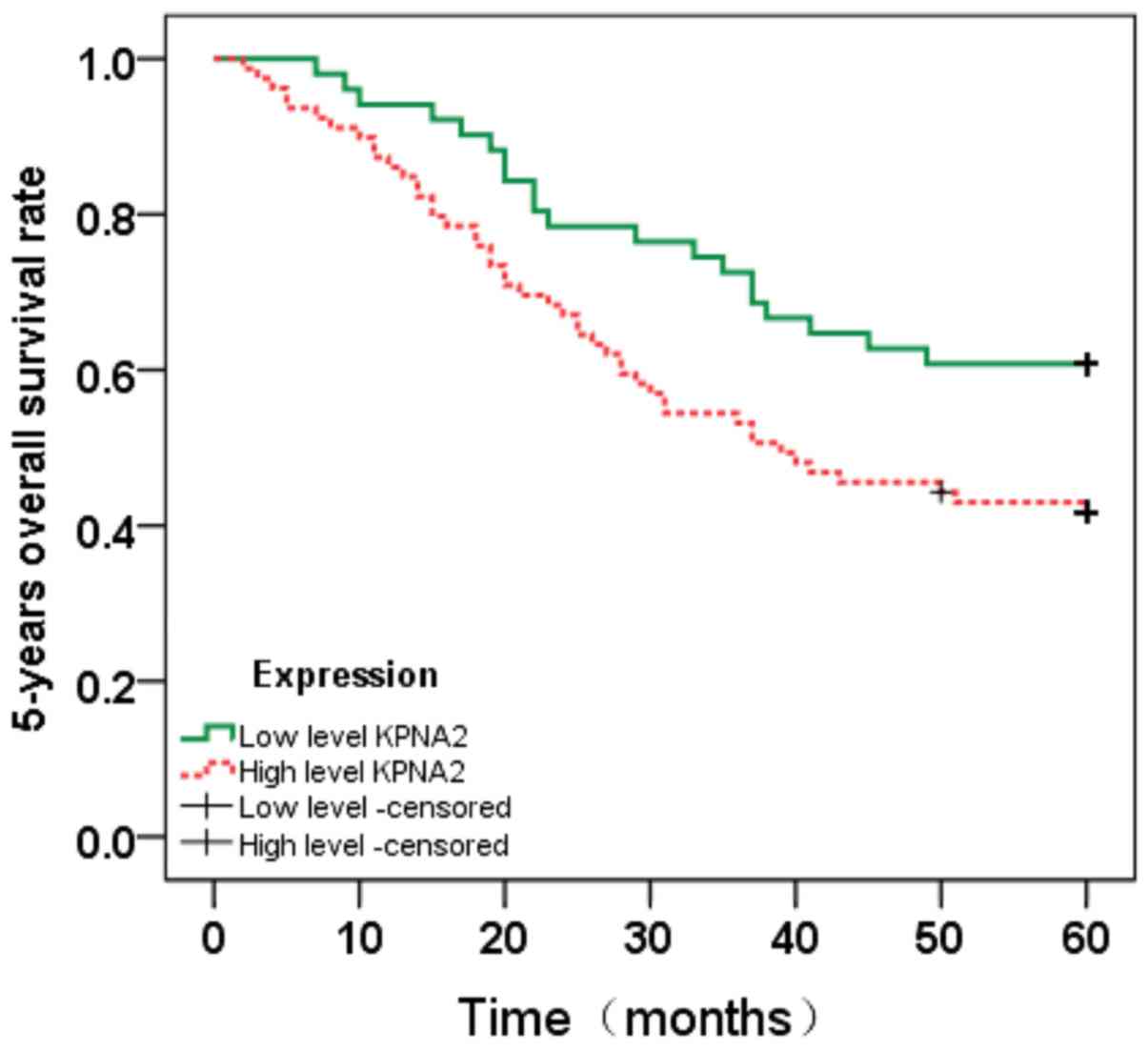
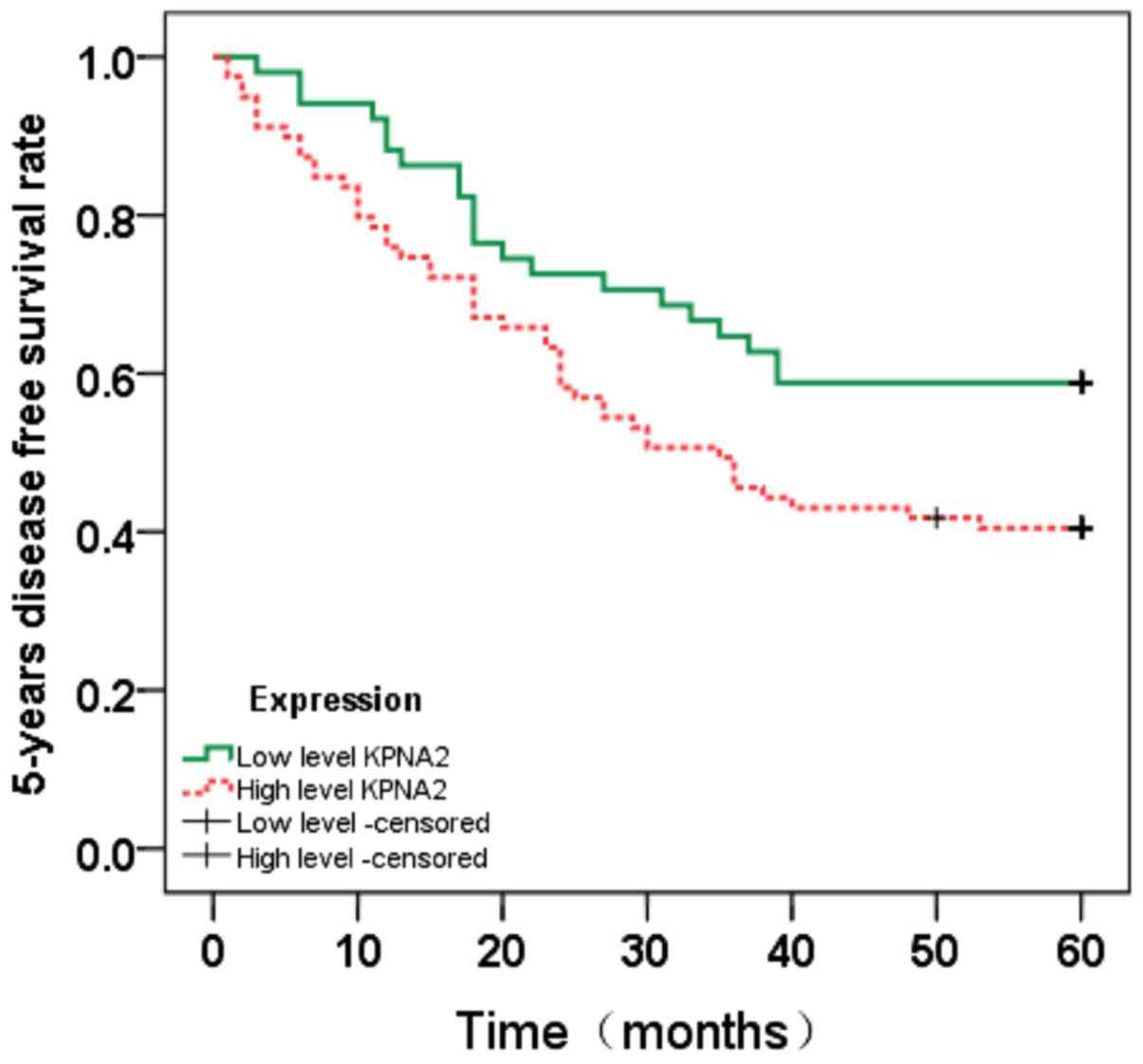
Survival analysis
To evaluate the prognostic value of KPNA2 for CRC,
the association between KPNA2 expression and patient survival time
was analyzed using the Kaplan-Meier method with a log-rank test
(Figs. 4 and 5). Nuclear KPNA2 expression was
significantly associated with overall survival (OS; P=0.026) and
disease-free survival (DFS; P=0.037). Overall, higher KPNA2
expression was associated with shorter OS and DFS times in CRC
patients.
Discussion
The present study revealed that KPNA2 expression is
upregulated in primary tumors and the serum of CRC patients. The
level of KPNA2 expression was associated with certain
clinicopathological features, including lymph node involvement,
LVI, PNI, tumor differentiation, infiltration depth and TNM stage.
Furthermore, high levels of KPNA2 expression were observed to
correlate with shorter survival times in CRC patients following
radical surgery.
The exchange of molecules between the nucleus and
cytoplasm is mediated by large nuclear pore complexes (2). Karyopherins are involved in this process
when molecules are >40 kDa (8),
acting as carrier proteins in a selective bidirectional shuttling
process (8). In cancer cells,
cellular transport dysfunction is common (8).
In 2006, Dahl et al (9) reported that KPNA2 expression exhibited
prognostic value in breast cancer, and this was subsequently
demonstrated in a variety of other malignancies, including prostate
and gastric cancer (10–18). In recent years, KPNA2 has gained
attention as a potential biomarker for several types of cancer,
including lung cancer. The increased KPNA2 expression observed in
cancer tissues is predominantly localized to the cell nucleus. This
may be due to cellular stress, including oxidative stress and heat
shock, which may lead to the accumulation of KPNA2 in the nucleus
of tumor cells. It has been reported that cells in advanced tumors
typically exhibit a high level of oxidative stress (19–25).
KPNA2 overexpression in cancer tissues has been
associated with increased morbidity (7,13). The
majority of previous studies have shown that increased KPNA2
expression is correlated with a poor prognosis (7,9,13). Other studies have implicated KPNA2 as
an independent prognostic factor for patients with breast and lung
cancer (9,15–17).
Similarly, Kaplan-Meier survival analysis performed in the present
study revealed that KPNA2 expression in primary tumors is a
powerful predictive factor for CRC patients. Therefore, these
results indicate that measuring KPNA2 expression may allow
physicians to diagnose CRC patients with a poor prognosis. This may
lead to better individualized risk stratification and thus, may
optimize therapy for CRC patients.
Due to its involvement in molecular transport and as
a tumor marker, KPNA2 has a role in a number of biological
processes, including cellular proliferation, differentiation,
cellular matrix adhesion, colony formation and migration (4,13,15,26–28).
Recent evidence has indicated that KPNA2 may regulate cancer cell
transformation (26). KPNA2
expression in cancer tissues is typically 5–10-fold higher than
that in normal tissue at the transcriptional level (10,14). These
differences in KPNA2 expression were additionally observed in the
present study, which supports the hypothesis that KPNA2 possesses a
significant role in CRC carcinogenesis. Certain viruses cause their
host cells to proliferate uncontrollably (28). Such viruses, often termed tumor
viruses, may lead to carcinogenesis (28). Notably, a previous study revealed that
human papilloma virus, BK virus and Simian virus 40 are capable of
inducing chromosomal instability and contributing to CRC
development by altering cell cycle control and inhibiting apoptosis
(29). KPNA2 has been reported to be
involved in tumor viral infections, primarily by transporting
viruses, including Epstein-Barr virus (30), human papillomavirus (31,32),
polyomavirus (33) and human
immunodeficiency virus (34), into
the nucleus. These previous studies (30–34)
demonstrated that viruses infected the normal host cells and led to
cell carcinogenesis via the KPNA2 signaling pathway (27). Furthermore, KPNA2 is hypothesized to
function in the regulation of viral capsid assembly (35). It has additionally been postulated
that KPNA2 may import DNA repair proteins and cell cycle control
proteins, including breast cancer 1 and Nijmegen breakage syndrome
1 (36–38). In addition, KPNA2 has been implicated
in the cellular processes of the carcinogenesis of various types of
cancer (38). KPNA2 expression has
been associated with numerous differentiation processes. In
embryonic stem cells obtained from mice, due to promoter activation
by Krüppel-like factor (Klf)2 and Klf4, high expression of KPNA2
was observed (39). Silencing of
KPNA2 has been shown to suppress the migration and proliferation of
lung cancer cells, indicating the significance of KPNA2 in
tumorigenesis (13).
The present study revealed that KPNA2 expression is
associated with CRC tumor stage. Furthermore, it has been
demonstrated that KPNA2 expression is positively correlated with
lymph node metastasis and venous infiltration (9,11,14–17,40–42).
A previous study suggested that KPNA2 exhibits a significant role
in the malignant transformation of cancer cells (27). Thus, KPNA2 may serve as a novel
diagnostic factor for early stage cancer.
In conclusion, the present study revealed that KPNA2
is overexpressed during the malignant tumor progression of CRC. In
addition, univariate analyses demonstrated that increased KPNA2
expression was associated with shortened survival times in CRC
patients that had undergone radical surgery. Further molecular
biological experiments are required to investigate the mechanism by
which high levels of KPNA2 expression promote the progression of
CRC.
Acknowledgements
This paper was supported by the Foundation of
Heilongjiang Academy of Medical Science (grant no. 201601).
References
|
1
|
Brenner H, Kloor M and Pox CP: Colorectal
cancer. Lancet. 65:1490–1502. 2014. View Article : Google Scholar
|
|
2
|
Kau TR, Way JC and Silver PA: Nuclear
transport and cancer: From mechanism to intervention. Nat Rev
Cancer. 4:106–117. 2004. View
Article : Google Scholar : PubMed/NCBI
|
|
3
|
Goldfarb DS, Corbett AH, Mason DA,
Harreman MT and Adam SA: Importin alpha: A multipurpose
nuclear-transport receptor. Trends Cell Biol. 14:505–514. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Christiansen A and Dyrskjøt L: The
functional role of the novel biomarker karyopherin α 2 (KPNA2) in
cancer. Cancer Lett. 331:18–23. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Radu A, Blobel G and Moore MS:
Identification of a protein complex that is required for nuclear
protein import and mediates docking of import substrate to distinct
nucleoporins. Proc Natl Acad Sci USA. 92:1769–1773. 1995.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kotera I, Sekimoto T, Miyamoto Y, Saiwaki
T, Nagoshi E, Sakagami H, Kondo H and Yoneda Y: Importin alpha
transports CaMKIV to the nucleus without utilizing importin beta.
EMBO J. 24:942–951. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Stewart M: Molecular mechanism of the
nuclear protein import cycle. Nat Rev Mol Cell Biol. 8:195–208.
2007. View
Article : Google Scholar : PubMed/NCBI
|
|
8
|
Livak and Schmittgen, . Analysis of
relative gene expression data using real-time quantitative PCR and
the 2-ΔΔCt method. Methods. 25:402–408. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Dahl E, Kristiansen G, Gottlob K, Klaman
I, et al: Molecular profiling of laser-microdissected matched tumor
and normal breast tissue identifies karyopherin alpha2 as a
potential novel prognostic marker in breast cancer. Clin Cancer
Res. 12:3950–3960. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
van der Watt PJ, Maske CP, Hendricks DT,
Parker MI, Denny L, Govender D, Birrer MJ and Leaner VD: The
Karyopherin proteins, Crm1 and Karyopherin beta1, are overexpressed
in cervical cancer and are critical for cancer cell survival and
proliferation. Int J Cancer. 124:1829–1840. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Sakai M, Sohda M, Miyazaki T, Suzuki S,
Sano A, Tanaka N, Inose T, Nakajima M, Kato H and Kuwano H:
Significance of karyopherin-{alpha} 2 (KPNA2) expression in
esophageal squamous cell carcinoma. Anticancer Res. 30:851–856.
2010.PubMed/NCBI
|
|
12
|
Winnepenninckx V, Lazar V, Michiels S,
Dessen P, Stas M, Alonso SR, Avril MF, Romero PL Ortiz, Robert T,
Balacescu O, et al: Gene expression profiling of primary cutaneous
melanoma and clinical outcome. J Natl Cancer Inst. 98:472–482.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wang CI, Wang CL, Wang CW, Chen CD, Wu CC,
Liang Y, Tsai YH, Chang YS, Yu JS and Yu CJ: Importin subunit
alpha-2 is identified as a potential biomarker for non-small cell
lung cancer by integration of the cancer cell secretome and tissue
transcriptome. Int J Cancer. 128:2364–2372. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Li C, Ji L, Ding ZY, Zhang QD and Huang
GR: Overexpression of KPNA2 correlates with poor prognosis in
patients with gastric adenocarcinoma. Tumour Biol. 34:1021–1026.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Mortezavi A, Hermanns T, Seifert HH, et
al: KPNA2 expression is an independent adverse predictor of
biochemical recurrence after radical prostatectomy. Clin Cancer
Res. 17:1111–1121. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Gousias K, Becker AJ, Simon M and
Niehusmann P: Nuclear karyopherin a2: A novel biomarker for
infiltrative astrocytomas. J Neurooncol. 109:545–553. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yoshitake K, Tanaka S, Mogushi K, et al:
Importin-α1 as a novel prognostic target for hepatocellular
carcinoma. Ann Surg Oncol. 18:2093–2103. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
He L, Ding H, Wang JH, et al:
Overexpression of karyopherin 2 in human ovarian malignant germ
cell tumor correlates with poor prognosis. PLoS One. 7:e429922012.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Stochaj U, Rassadi R and Chiu J:
Stress-mediated inhibition of the classical nuclear protein import
pathway and nuclear accumulation of the small GTPase Gsp1p. FASEB
J. 14:2130–2132. 2000.PubMed/NCBI
|
|
20
|
Furuta M, Kose S, Koike M, et al:
Heat-shock induced nuclear retention and recycling inhibition of
importin alpha. Genes Cells. 9:429–441. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kodiha M, Chu A, Matusiewicz N and Stochaj
U: Multiple mechanisms promote the inhibition of classical nuclear
import upon exposure to severe oxidative stress. Cell Death Differ.
11:862–874. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Miyamoto Y, Saiwaki T, Yamashita J, Yasuda
Y, Kotera I, Shibata S, Shigeta M, Hiraoka Y, Haraguchi T and
Yoneda Y: Cellular stresses induce the nuclear accumulation of
importin alpha and cause a conventional nuclear import block. J
Cell Biol. 165:617–623. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Trachootham D, Alexandre J and Huang P:
Targeting cancer cells by ROS-mediated mechanisms: A radical
therapeutic approach? Nat Rev Drug Discov. 8:579–591. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yasuda Y, Miyamoto Y, Yamashiro T, Asally
M, Masui A, Wong C, Loveland KL and Yoneda Y: Nuclear retention of
importin α coordinates cell fate through changes in gene
expression. EMBO J. 31:83–94. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Miyamoto Y, Loveland KL and Yoneda Y:
Nuclear importin α and its physiological importance. Commun Integr
Biol. 5:220–222. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Umegaki N, Tamai K, Nakano H, Moritsugu R,
Yamazaki T, Hanada K, Katayama I and Kaneda Y: Differential
regulation of karyopherin alpha 2 expression by TGF-beta1 and
IFN-gamma in normal human epidermal keratinocytes: Evident
contribution of KPNA2 for nuclear translocation of IRF-1. J Invest
Dermatol. 127:1456–1464. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Noetzel E, Rose M, Bornemann J, Gajewski
M, Knüchel R and Dahl E: Nuclear transport receptor karyopherin-α2
promotes malignant breast cancer phenotypes in vitro. Oncogene.
31:2101–2114. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Hall MN, Griffin CA, Simionescu A, Corbett
AH and Pavlath GK: Distinct roles for classical nuclear import
receptors in the growth of multinucleated muscle cells. Dev Biol.
357:248–258. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Giuliani L, Ronci C, Bonifacio D, Di
Bonito L, Favalli C, Perno CF, Syrjänen K and Ciotti M: Detection
of oncogenic DNA viruses in colorectal cancer. Anticancer Res.
28:1405–1410. 2008.PubMed/NCBI
|
|
30
|
Fischer N, Kremmer E, Lautscham G,
Mueller-Lantzsch N and Grässer FA: Epstein-Barr virus nuclear
antigen 1 forms a complex with the nuclear transporter karyopherin
alpha2. J Biol Chem. 272:3999–4005. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Nelson LM, Rose RC, LeRoux L, Lane C,
Bruya K and Moroianu J: Nuclear import and DNA binding of human
papillomavirus type 45 L1 capsid protein. J Cell Biochem.
79:225–238. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Le Roux LG and Moroianu J: Nuclear entry
of high-risk human papillomavirus type 16 E6 oncoprotein occurs via
several pathways. J Virol. 77:2330–2337. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Qu Q, Sawa H, Suzuki T, Semba S, Henmi C,
Okada Y, Tsuda M, Tanaka S, Atwood WJ and Nagashima K: Nuclear
entry mechanism of the human polyomavirus JC virus-like particle:
Role of importins and the nuclear pore complex. J Biol Chem.
279:27735–27742. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Gallay P, Stitt V, Mundy C, Oettinger M
and Trono D: Role of the karyopherin pathway in human
immunodeficiency virus type 1 nuclear import. J Virol.
70:1027–1032. 1996.PubMed/NCBI
|
|
35
|
Bird G, O'Donnell M, Moroianu J and Garcea
RL: Possible role for cellular karyopherins in regulating
polyomavirus and papillomavirus capsid assembly. J Virol.
82:9848–9857. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Narod SA and Foulkes WD: BRCA1 and BRCA2:
1994 and beyond. Nat Rev Cancer. 4:665–676. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Teng SC, Wu KJ, Tseng SF, Wong CW and Kao
L: Importin KPNA2, NBS1, DNA repair and tumorigenesis. J Mol
Histol. 37:293–299. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Cutress ML, Whitaker HC, Mills IG, Stewart
M and Neal DE: Structural basis for the nuclear import of the human
androgen receptor. J Cell Sci. 121:957–968. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Kamikawa Y, Yasuhara N and Yoneda Y: Cell
type-specific transcriptional regulation of the gene encoding
importin-alpha1. Exp Cell Res. 317:1970–1978. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Dankof A, Fritzsche FR, Dahl E, Pahl S,
Wild P, Dietel M, Hartmann A and Kristiansen G: KPNA2 protein
expression in invasive breast carcinoma and matched peritumoral
ductal carcinoma in situ. Virchows Arch. 451:877–881. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Gluz O, Wild P, Meiler R, Diallo-Danebrock
R, Ting E, Mohrmann S, Schuett G, Dahl E, Fuchs T, Herr A, et al:
Nuclear karyopherin alpha2 expression predicts poor survival in
patients with advanced breast cancer irrespective of treatment
intensity. Int J Cancer. 123:1433–1438. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Altan B, Yokobori T, Mochiki E, Ohno T,
Ogata K, Ogawa A, Yanai M, Kobayashi T, Luvsandagva B, Asao T and
Kuwano H: Nuclear karyopherin-α2 expression in primary lesions and
metastatic lymph nodes was associated with poor prognosis and
progression in gastric cancer. Carcinogenesis. 34:2314–2321. 2013.
View Article : Google Scholar : PubMed/NCBI
|