In the early 20th century, chromosomal abnormalities
were considered to cause carcinogenesis (1). The subsequent identification of various
oncogenes and tumor-suppressor genes demonstrated that cancer
develops due to genetic abnormalities (2,3); however,
single-gene abnormalities are insufficient to explain the malignant
transformation of all types of cancer, which are hypothesized to
require a combination of numerous genetic abnormalities (4). To investigate the underlying mechanisms
of cancer development, a human genome-wide analysis study was
proposed in 1986, and the Human Genome Project was conducted from
1990 to 2003 (5). Genome-wide
analyses of various types of cancer have subsequently increased due
to the widespread use of next-generation sequencers (6).
Genome-wide analyses have revealed that B-rapidly
accelerated fibrosarcoma proto-oncogene, serine/threonine kinase
(BRAF) and phosphatidylinositol-4,5-bisphosphate 3-kinase,
catalytic subunit α (PIK3CA) are frequently expressed in
melanoma and rectal cancer, respectively (7,8), and that
epidermal growth factor receptor genetic abnormalities may occur in
lung cancers (9–11). The Cancer Genome Atlas (CGA) project
was conducted in the USA and has collected data on 12 types of
cancer (cervical cancer, cholangiocarcinoma, esophageal carcinoma,
liver hepatocellular carcinoma, mesothelioma, pancreatic ductal
adenocarcinoma, paraganglioma and pheochromocytoma, sarcoma,
testicular germ cell cancer, thymoma, uterine carcinosarcoma and
uveal melanoma) since 2006 till 2012. The objective of the CGA was
to conduct comprehensive analyses of complete genomes for various
types of cancer and to compare them with normal human genomes, with
the aim of identifying the presence of genetic abnormalities in
each type of cancer (12). The CGA
aims to conduct further analyses in other types of cancer in order
to determine characteristic and common genetic abnormalities among
various types of cancer (13).
Data from the tumor genomes of several hundred
patients with ovarian cancer, brain tumors or squamous cell lung
cancer, and comparative data from normal genomes, have been
analyzed in the CGA (14–16). These results were published in 2009,
and formed the basis for the Pan-Cancer Analysis Project, which
began in 2012 (17). In that study on
5,074 patients, the exomes of 12 types of cancer were analyzed,
including bladder urothelial carcinoma, breast and colon cancers,
head and neck squamous cell carcinoma, renal clear cell carcinoma,
acute myeloid leukemia, lung adenocarcinoma, squamous cell lung
cancer, and ovarian, rectal and endometrial cancer (17). Comprehensive genome-wide analysis was
used to identify common or independent molecular characteristics
among these types of cancer (18).
Regarding gynecologic cancers, all exomes of ovarian clear cell
carcinoma were analyzed in 2010, and the results of the genomic
analyses of ovarian serous carcinoma and endometrial cancer were
published in 2011 and 2013, respectively (19,20).
Ovarian clear cell carcinoma accounts for 8.4% of
all ovarian cancers worldwide (21).
Mutations in the PIK3CA gene were identified to occur in
~33% of cases of ovarian clear cell carcinoma (22). Tan et al identified protein
kinase B 2 (AKT2) gene amplifications in certain clear cell
carcinomas, and demonstrated its involvement in patient prognosis
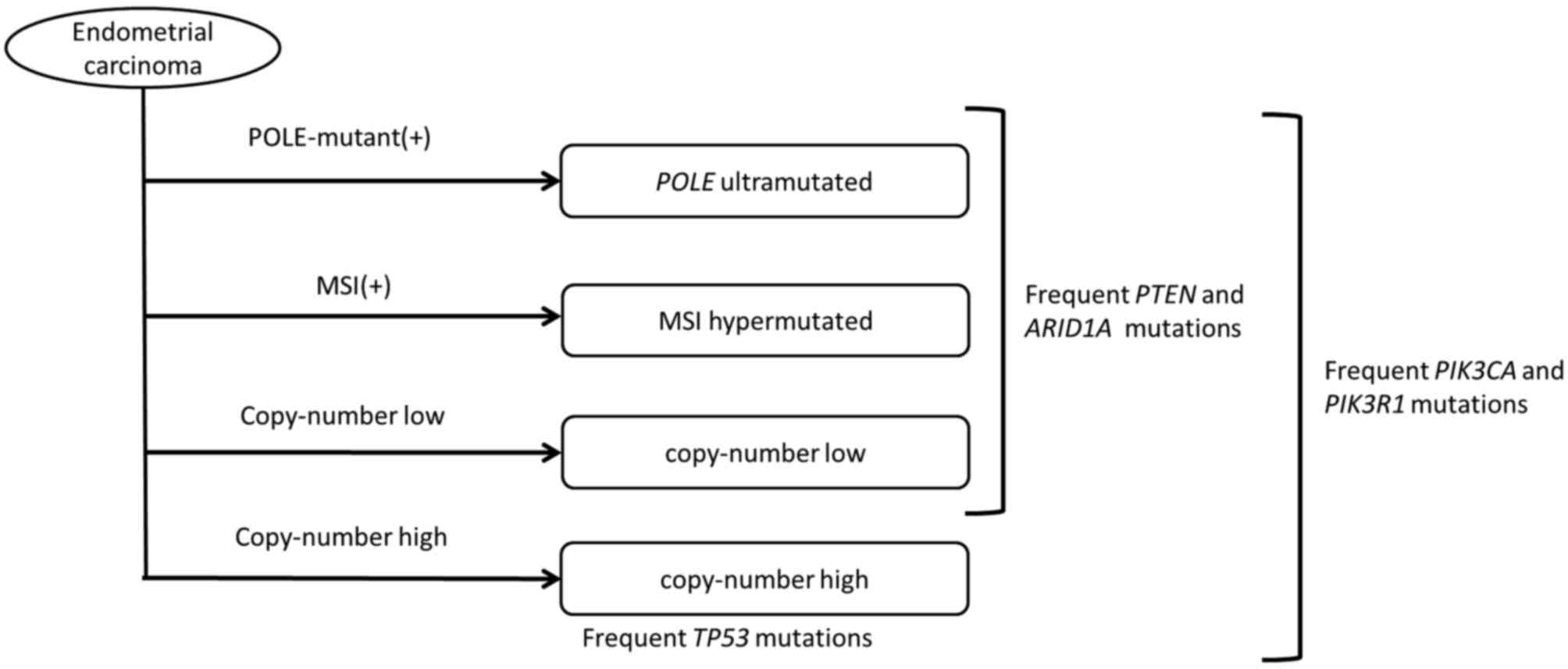
(23). The CGA identified a number of
frequent mutations in the AT-rich interactive domain 1A
(ARID1A) gene, which is located on chromosome 1q36 and
encodes Brahma/SWI2-related gene 1-associated factor 250 (24). The transcription product of the
ARIDIA gene is a component of the SWItch/sucrose
non-fermentable chromatin remodeling complex (25), which alters the nucleosomal structure
and regulates DNA-binding proteins in an adenosine
triphosphate-dependent manner (26).
Therefore, abnormalities in these complexes may cause abnormalities
in DNA transcription, replication and repair, and may result in the
malignant transformation of cells. Abnormalities in this specific
component protein have been frequently identified in patients with
ovarian clear cell carcinoma, particularly at International
Federation of Gynecology and Obstetrics stages III and IV, and in
patients with high cancer antigen 125 expression levels (27).
In order to examine the messenger RNA (mRNA)
expression pattern in clear cell carcinoma, ~400 gene groups with
differential expression profiles specific to clear cell carcinoma
were selected, and the signature of ovarian clear cell carcinoma
was identified (28). Mutations in
hepatocyte nuclear factor-1β (29)
and ARID1A (24) are currently
considered to be important for occurrence of clear cell carcinoma,
and numerous genes that are associated with the ovarian clear cell
carcinoma signature are involved in stress response, glucose
metabolism and coagulation, which are three key signaling pathways
in clear cell carcinoma (30).
Numerous patients with clear cell adenocarcinoma originally present
with endometriosis, which suggests that the microenvironment in
endometriosis includes signaling factors that may be involved in
the development of this type of cancer (28). In a typical case of endometriosis,
high levels of free iron are observed, which may generate reactive
oxygen species (31). In addition,
the oxidative stress levels are high, and cell dysfunction and DNA
damage are common (31). Stress
response genes are often highly expressed in patients with clear
cell adenocarcinoma who originally had endometriosis, which
indicates that the stress response signaling pathway may be
involved in the development of clear cell carcinoma from
endometriosis (28).
In serous ovarian cancer, the incidence of mutation
is also frequent (~22%) for the breast cancer 1/2 (BRCA1/2)
gene (39). BRCA is involved
in homologous recombination and thus, mutations in this gene may
cause defects in DNA repair mechanisms (40). Abnormal homologous recombination has
been identified in ~50% of cells with BRCA mutations
(41). Mutations in BRCA2-interacting
transcriptional repressor (EMSY; 8%), phosphatase and tensin
homolog (PTEN; 7%), RAD51 paralog C (3%) and Fanconi anemia
complementation group D2 (5%) have also been identified in various
serous carcinomas (19).
A DNA copy number analysis was also performed in the
CGA, as chromosomal instability is a characteristic of high-grade
serous carcinoma (42). High-grade
serous carcinoma has more changes in DNA copy number than other
tissue types of epithelial ovarian cancer (43). DNA copy number abnormalities in HGSOC
include amplification of the v-myc avian myelocytomatosis viral
oncogene homolog (MYC) and cyclin E1 (CCNE1) genes,
as well as BRCA defects, a number of which are considered to
be involved in patient prognosis (44). The CGA analysis identified complex DNA
copy number abnormalities in >1/2 of the patients involved
(19), with chromosomal regions
including CCNE1, MYC and MDS1 and EVI1 complex locus
protein EVI1 being highly amplified in >20% of tumors (45). Five specific amplified genes were
identified, including the activated C-kinase receptor zinc finger
MYND-type containing 8, the p53 target gene interferon regulatory
factor 2 binding protein 2, the DNA-binding protein inhibitor
inhibitor of differentiation 4, the embryonic development gene
paired box 8 and the telomerase catalytic subunit telomerase
reverse transcriptase (TERT) (19).
The results of the analysis described above indicate
that >20 gene abnormalities may occur in high-grade serous
adenocarcinoma, and that these are often identified in certain
signaling molecules, including retinoblastoma, PI3K/AKT, neurogenic
locus notch homolog and FOXM1 (46).
Analyses of ovarian and endometrial cancers reveal
common gene abnormalities, despite tumor development in various
tissues, whereas gene abnormalities may differ in certain types of
cancer that develop in the same tissues (Tables I and II) (70). In
ovarian cancer, TP53 mutations occur in 96% of high-grade
serous adenocarcinoma cases (71).
HGSOC has a poor prognosis (63),
which previously led to the proposal to classify ovarian cancer
according to the type of gene mutation, rather than the tissue type
(71). In the proposed
classification, type I includes low-grade serous adenocarcinoma,
low-grade endometrioid carcinoma, clear cell carcinoma and serous
carcinoma, which have infrequent TP53 mutations, while type
II includes high-grade serous carcinoma, undifferentiated cancer
and carcinosarcoma, which have frequent TP53 mutations
(72). The classification according
to the frequency of TP53 mutations facilitates screening and
therapy (73), and may allow the
development of personalized treatments.
Novel treatments targeting specific gene
abnormalities have been developed based on the CGA results. In a
previous study, 6–12 cycles of platinum-based adjuvant chemotherapy
were administered following tumor debulking in 60 patients with
ovarian clear cell adenocarcinoma and 17 patients with high-grade
serous adenocarcinoma (27). An
evaluation of prognosis and ARID1A expression levels demonstrated
that the PFS in the ARID1A-negative group was significantly
shorter, as compared with that in the ARID1A-positive group
(P<0.01), indicating a resistance to platinum-based chemotherapy
in the absence of ARID1A (27).
Therefore, the results of genome-wide analysis revealed the
mechanisms underlying the variation in chemoresistance among these
patients. Current chemotherapeutic strategies for ovarian cancer
are determined based on the disease stage; however, conventional
chemotherapy may be disadvantageous for certain patients, and
treatment may be more effectively selected based on the presence of
specific gene mutations.
Poly (ADP-ribose) polymerase (PARP) inhibitors for
BRCA1/2 are currently being developed as novel anticancer drugs
targeting tumors with gene abnormalities (76). PARP is activated following DNA damage,
and repairs single-stranded DNA by polymerization of adenosine
diphosphate-ribose residues (77). In
the absence of DNA repair by PARP, double-stranded DNA is repaired
by BRCA1/2 (78). In tumors with
BRCA1/2 mutations, the use of PARP inhibitors eliminates all
DNA repair mechanisms, inducing an antitumor effect via the
promotion of cell death (79,80). The CGA identified numerous
BRCA1/2 mutations in HGSOC, and olaparib, a PARP inhibitor,
has been examined in a phase II clinical trial in patients with
this disease (81). Trials using a
combination of current chemotherapeutic agents for ovarian cancer
and olaparib are also in progress (82,83). Thus,
the treatment of ovarian cancer may improve due to the enhanced
understanding of the genetic abnormalities involved in this
disease.
PI3K/mechanistic target of rapamycin (mTOR)
inhibition may be effective for the treatment of HGSOC and
endometrial cancer with mutations in the PI3K/AKT signaling
pathway. mTOR is a serine/threonine kinase that regulates cell
proliferation (84). Certain gene
mutations increase mTOR expression levels, resulting in abnormal
cell proliferation (84).
Hypoxia-inducible factor 1α is located downstream of mTOR, and is
involved in angiogenesis; thus, mTOR overexpression enhances cell
proliferation and vasa vasorum neovascularization (85–87).
Anticancer drugs targeting mTOR include everolimus, an mTOR
inhibitor used to treat renal cell carcinoma (88). Gene analysis of ovarian cancer tissues
revealed mTOR mutations, therefore suggesting the effectiveness of
mTOR inhibitors in ovarian cancer (88–90). In
mice with subcutaneously implanted human ovarian clear cell
carcinoma cells, the tumor size was halved following treatment with
everolimus, Taxol® and cisplatin (86). When everolimus was administered alone,
the tumor size did not decrease, but cellular apoptosis was
observed (86).
As previously described, DNA copy number
abnormalities occur at a high rate in type II HGSOC, with
amplification of ≥22 oncogenes (19).
Therefore, specific gene abnormalities are present in numerous
signaling pathways in high-grade serous adenocarcinoma (91). A molecular-targeted drug may only
inhibit one signaling pathway and thus, conventional chemotherapy
may be required to obtain a good therapeutic effect (92). Trials of novel drugs that are able to
simultaneously inhibit PI3K and mTOR have also been conducted
(93,94). These include a phase I study on the
use of DS-7423 for the treatment of ovarian cancer and a phase I
study on the use of NVP-BEZ235 for the treatment of endometrial
cancer, in which, the efficacy of NVP-BEZ235 was compared with that
of everolimus (94).
Ovarian low-grade serous carcinoma also has poor
sensitivity to standard chemotherapy (95,96). In
this disease, KRAS and BRAF are important in the MAPK signaling
pathways, and the MAPK kinase inhibitor selumetinib has an
antitumor effect (97,98). A phase II study demonstrated a
response rate of 15.4%, a disease progression-control rate of 80.8%
and a median PFS of 11.0 months (98). In contrast to HGSOC, the low-grade
type of serous ovarian carcinoma has relatively lower DNA copy
number, and the underlying mechanism of carcinogenesis may depend
on a single signaling pathway (99).
Therefore, the efficacy of specific molecular-targeted drugs is
consistent with the CGA analysis.
CGA analyses of gene mutations have provided novel
classifications and the foundation for therapy selection based on
gene abnormalities. Epigenetic mutations include those in microRNAs
(miRNAs or miRs) (100–102), which are small RNA molecules
containing ~22 nucleotides that induce gene silencing (102,103).
In gynecologic cancers, the downregulation of specific miRNAs,
including miR-30c and miR-152, is involved in the onset of cancer
(104,105). These miRNAs downregulate the
expression of certain oncogenes, including discoidin domain
receptor and latent transforming growth factor-β binding protein-4;
therefore, decreased expression of these miRNAs results in
tumorigenesis (106). Further
analyses of epigenetic changes, including those associated with
miRNAs, are required for comparison with the analyses of gene
mutations.
The causes of gene mutations also require further
evaluation. The CGA results indicate that the apolipoprotein B mRNA
editing enzyme catalytic polypeptide-like 3B (APOBEC3B) is a cause
of certain gene mutations (107–109),
with a C-T base substitution rate identified to be more frequent
than other substitutions in numerous types of cancer, including
bladder and lung cancer (110).
APOBEC3B is overexpressed in cancer cells with frequent C-T base
substitutions, which suggests that APOBEC3B may be associated with
genome mutations (108).
The genome-wide analyses in the CGA project have
detected common abnormalities in various types of cancer (109). However, only 12 types of cancer have
been analyzed to date, and the CGA aims to analyze a further 50
types of cancer through extended data collection (109). Genome-wide analyses were initially
intended to determine the mechanisms underlying the development of
cancer; however, the pathogenic mechanisms have yet to be
elucidated, despite the important information previously obtained
on gene abnormalities (17).
Epigenetic data, including those for miRNAs, and proteomic analyses
have revealed that carcinogenesis depends on numerous other factors
in addition to gene abnormalities (110). These results indicate that further
understanding of the pathogenic mechanisms underlying cancer will
require numerous genomic, epigenetic, transcriptomic and proteomic
studies, including current genome, miRNA expression, DNA
methylation and reverse-phase protein array analyses.
The authors thank Dr S. Takizawa (Keio University
School of Medicine, Tokyo, Japan) and Dr A. Chida (Keio University
School of Medicine, Tokyo, Japan) for their helpful assistance. The
present study was supported by the Keio Gijuku Academic Development
Fund (Tokyo, Japan).
|
1
|
Boveri T: On multipolar mitosis as a means
of analysis of the cell nucleus. Neu Folge. 35:67–90. 1902.
|
|
2
|
Stehelin D, Varmus HE, Bishop JM and Vogt
PK: DNA related to the transforming gene(s) of avian sarcoma
viruses is present in normal avian DNA. Nature. 260:170–173. 1976.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Tabin CJ, Bradley SM, Bargmann CI,
Weinberg RA, Papageorge AG, Scolnick EM, Dhar R, Lowy DR and Chang
EH: Mechanism of activation of a human oncogene. Nature.
300:143–149. 1982. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Vogelstein B, Fearon ER, Hamilton SR, Kern
SE, Preisinger AC, Leppert M, Nakamura Y, White R, Smits AM and Bos
JL: Genetic alterations during colorectal-tumor development. N Engl
J Med. 319:525–532. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Dulbecco R: A turning point in cancer
research: Sequencing the human genome. Science. 231:1055–1056.
1986. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wright CF, Fitzgerald TW, Jones WD,
Clayton S, McRae JF, van Kogelenberg M, King DA, Ambridge K,
Barrett DM, Bayzetinova T, et al: Genetic diagnosis of
developmental disorders in the DDD study: A scalable analysis of
genome-wide research data. Lancet. 385:1305–1314. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Davies H, Bignell GR, Cox C, Stephens P,
Edkins S, Clegg S, Teague J, Woffendin H, Garnett MJ, Bottomley W,
et al: Mutations of the BRAF gene in human cancer. Nature.
417:949–954. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Samuels Y, Wang Z, Bardelli A, Silliman N,
Ptak J, Szabo S, Yan H, Gazdar A, Powell SM, Riggins GJ, et al:
High frequency of mutations of the PIK3CA gene in human cancers.
Science. 304:5542004. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lynch TJ, Bell DW, Sordella R,
Gurubhagavatula S, Okimoto RA, Brannigan BW, Harris PL, Haserlat
SM, Supko JG, Haluska FG, et al: Activating mutations in the
epidermal growth factor receptor underlying responsiveness of
non-small-cell lung cancer to gefitinib. N Engl J Med.
350:2129–2139. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Paez JG, Jänne PA, Lee JC, Tracy S,
Greulich H, Gabriel S, Herman P, Kaye FJ, Lindeman N, Boggon TJ, et
al: EGFR mutations in lung cancer: Correlation with clinical
response to gefitinib therapy. Science. 304:1497–1500. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Pao W, Miller V, Zakowski M, Doherty J,
Politi K, Sarkaria I, Singh B, Heelan R, Rusch V, Fulton L, et al:
EGF receptor gene mutations are common in lung cancers from ‘never
smokers’ and are associated with sensitivity of tumors to gefitinib
and erlotinib. Proc Natl Acad Sci USA. 101:13306–13311. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Akbani R, Ng PK, Werner HM, Shahmoradgoli
M, Zhang F, Ju Z, Liu W, Yang JY, Yoshihara K, Li J, et al: A
pan-cancer proteomic perspective on The Cancer Genome Atlas. Nat
Commun. 5:38872014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
The future of cancer genomics. Nat Med.
21:992015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Garraway LA and Lander ES: Lessons from
the cancer genome. Cell. 153:17–37. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Vogelstein B, Papadopoulos N, Velculescu
VE, Zhou S, Diaz LA Jr and Kinzler KW: Cancer genome landscapes.
Science. 339:1546–1558. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wheeler DA and Wang L: From human genome
to cancer genome: The first decade. Genome Res. 23:1054–1062. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Cancer Genome Atlas Research Network;
Weinstein JN, Collisson EA, Mills GB, Shaw KR, Ozenberger BA,
Ellrott K, Shmulevich I, Sander C and Stuart JM: The cancer genome
atlas pan-cancer analysis project. Nat Genet. 45:1113–1120. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kong J, Cooper LA, Wang F, Gutman DA, Gao
J, Chisolm C, Sharma A, Pan T, Van Meir EG, Kurc TM, et al:
Integrative, multimodal analysis of glioblastoma using TCGA
molecular data, pathology images and clinical outcomes. IEEE Trans
Biomed Eng. 58:3469–3474. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Cancer Genome Atlas Research Network, .
Integrated genomic analyses of ovarian carcinoma. Nature.
474:609–615. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Cancer Genome Atlas Research Network, .
Kandoth C, Schultz N, Cherniack AD, Akbani R, Liu Y, Shen H,
Robertson AG, Pashtan I, Shen R, et al: Integrated genomic
characterization of endometrial carcinoma. Nature. 497:67–73. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Heintz AP, Odicino F, Maisonneuve P, Quinn
MA, Benedet JL, Creasman WT, Ngan HY, Pecorelli S and Beller U:
Carcinoma of the ovary. FIGO 26th annual report on the results of
treatment in gynecological cancer. Int J Gynaecol Obstet. 95:(Suppl
1). S161–S192. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kuo KT, Mao TL, Jones S, Veras E, Ayhan A,
Wang TL, Glas R, Slamon D, Velculescu VE, Kuman RJ and Shih Ie M:
Frequent activating mutations of PIK3CA in ovarian clear cell
carcinoma. Am J Pathol. 174:1597–1601. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Tan DS, Iravani M, McCluggage WG, Lambros
MB, Milanezi F, Mackay A, Gourley C, Geyer FC, Vatcheva R, Millar
J, et al: Genomic analysis reveals the molecular heterogeneity of
ovarian clear cell carcinomas. Clin Cancer Res. 17:1521–1534. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Jones S, Wang TL, Shih Ie M, Mao TL,
Nakayama K, Roden R, Glas R, Slamon D, Diaz LA Jr, Vogelstein B, et
al: Frequent mutations of chromatin remodeling gene ARID1A in
ovarian clear cell carcinoma. Science. 330:228–231. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wiegand KC, Shah SP, Al-Agha OM, Zhao Y,
Tse K, Zeng T, Senz J, McConechy MK, Anglesio MS, Kalloger SE, et
al: ARID1A mutations in endometriosis-associated ovarian
carcinomas. N Engl J Med. 363:1532–1543. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Mohrmann L and Verrijzer CP: Composition
and functional specificity of SWI2/SNF2 class chromatin remodeling
complexes. Biochim Biophys Acta. 1681:59–73. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Katagiri A, Nakayama K, Rahman MT, Rahman
M, Katagiri H, Nakayama N, Ishikawa M, Ishibashi T, Iida K,
Kobayashi H, et al: Loss of ARID1A expression is related to shorter
progression-free survival and chemoresistance in ovarian clear cell
carcinoma. Mod Pathol. 25:282–288. 2012.PubMed/NCBI
|
|
28
|
Yamaguchi K, Mandai M, Oura T, Matsumura
N, Hamanishi J, Baba T, Matsui S, Murphy SK and Konishi I:
Identification of an ovarian clear cell carcinoma gene signature
that reflects inherent disease biology and the carcinogenic
processes. Oncogene. 29:1741–1752. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Tsuchiya A, Sakamoto M, Yasuda J, Chuma M,
Ohta T, Ohki M, Yasugi T, Taketani Y and Hirohashi S: Expression
profiling in ovarian clear cell carcinoma: Identification of
hepatocyte nuclear factor-1 beta as a molecular marker and a
possible molecular target for therapy of ovarian clear cell
carcinoma. Am J Pathol. 163:2503–2512. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Yamaguchi K, Huang Z, Matsumura N, Mandai
M, Okamoto T, Baba T, Konishi I, Berchuck A and Murphy SK:
Epigenetic determinants of ovarian clear cell carcinoma biology.
Int J Cancer. 135:585–597. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Yamaguchi K, Mandai M, Toyokuni S,
Hamanishi J, Higuchi T, Takakura K and Fujii S: Contents of
endometriotic cysts, especially the high concentration of free
iron, are a possible cause of carcinogenesis in the cysts through
the iron-induced persistent oxidative stress. Clin Cancer Res.
14:32–40. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Barger CJ, Zhang W, Hillman J, Stablewski
AB, Higgins MJ, Vanderhyden BC, Odunsi K and Karpf AR: Genetic
determinants of FOXM1 overexpression in epithelial ovarian cancer
and functional contribution to cell cycle progression. Oncotarget.
6:27613–27627. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Barsotti AM and Prives C:
Pro-proliferative FOXM1 is a target of p53-mediated repression.
Oncogene. 28:4295–4305. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Ma RY, Tong TH, Cheung AM, Tsang AC, Leung
WY and Yao KM: Raf/MEK/MAPK signaling stimulates the nuclear
translocation and transactivating activity of FOXM1c. J Cell Sci.
118:795–806. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Tan Y, Raychaudhuri P and Costa RH: Chk2
mediates stabilization of the FOXM1 transcription factor to
stimulate expression of DNA repair genes. Mol Cell Biol.
27:1007–1016. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Lane DP: Cancer. p53, guardian of the
genome. Nature. 358:15–16. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Brachova P, Mueting SR, Carlson MJ,
Goodheart MJ, Button AM, Mott SL, Dai D, Thiel KW, Devor EJ and
Leslie KK: TP53 oncomorphic mutations predict resistance to
platinum- and taxane-based standard chemotherapy in patients
diagnosed with advanced serous ovarian carcinoma. Int J Oncol.
46:607–618. 2015.PubMed/NCBI
|
|
38
|
Brachova P, Mueting SR, Devor EJ and
Leslie KK: Oncomorphic TP53 mutations in gynecologic cancers lose
the normal protein: Protein interactions with the microRNA
microprocessing complex. J Cancer Ther. 5:506–516. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Liu G, Yang D, Sun Y, Shmulevich I, Xue F,
Sood AK and Zhang W: Differing clinical impact of BRCA1 and BRCA2
mutations in serous ovarian cancer. Pharmacogenomics. 13:1523–1535.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Farmer H, McCabe N, Lord CJ, Tutt AN,
Johnson DA, Richardson TB, Santarosa M, Dillon KJ, Hickson I,
Knights C, et al: Targeting the DNA repair defect in BRCA mutant
cells as a therapeutic strategy. Nature. 434:917–921. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Scully R, Puget N and Vlasakova K: DNA
polymerase stalling, sister chromatid recombination and the BRCA
genes. Oncogene. 19:6176–6183. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Kurman RJ and Shih Ie M: Molecular
pathogenesis and extraovarian origin of epithelial ovarian cancer:
Shifting the paradigm. Hum Pathol. 42:918–931. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Shah RH, Scott SN, Brannon AR, Levine DA,
Lin O and Berger MF: Comprehensive mutation profiling by
next-generation sequencing of effusion fluids from patients with
high-grade serous ovarian carcinoma. Cancer Cytopathol.
123:289–297. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Kurman RJ and Shih Ie M: Molecular
pathogenesis and extraovarian origin of epithelial ovarian
cancer-shifting the paradigm. Hum Pathol. 42:918–931. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Wiedemeyer WR, Beach JA and Karlan BY:
Reversing platinum resistance in high-grade serous ovarian
carcinoma: Targeting BRCA and the homologous recombination system.
Front Oncol. 4:342014. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Rao SS, O'Neil J, Liberator CD, Hardwick
JS, Dai X, Zhang T, Tyminski E, Yuan J, Kohl NE, Richon VM, et al:
Inhibition of NOTCH signaling by gamma secretase inhibitor engages
the RB pathway and elicits cell cycle exit in T-cell acute
lymphoblastic leukemia cells. Cancer Res. 69:3060–3068. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Jones S, Wang TL, Kurman RJ, Nakayama K,
Velculescu VE, Vogelstein B, Kinzler KW, Papadopoulos N and Shih Ie
M: Low-grade serous carcinomas of the ovary contain very few point
mutations. J Pathol. 226:413–420. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Tothill RW, Tinker AV, George J, Brown R,
Fox SB, Lade S, Johnson DS, Trivett MK, Etemadmoghadam D, Locandro
B, et al: Novel molecular subtypes of serous and endometrioid
ovarian cancer linked to clinical outcome. Clin Cancer Res.
14:5198–5208. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Rajagopalan H, Bardelli A, Lengauer C,
Kinzler KW, Vogelstein B and Velculescu VE: Tumorigenesis: RAF/RAS
oncogenes and mismatch-repair status. Nature. 418:9342002.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Singer G, Oldt R III, Cohen Y, Wang BG,
Sidransky D, Kurman RJ and Shih Ie M: Mutations in BRAF and KRAS
characterize the development of low-grade ovarian serous carcinoma.
J Natl Cancer Inst. 95:484–486. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Kannan K, Coarfa C, Chao PW, Luo L, Wang
Y, Brinegar AE, Hawkins SM, Milosavljevic A, Matzuk MM and Yen L:
Recurrent BCAM-AKT2 fusion gene leads to a constitutively activated
AKT2 fusion kinase in high-grade serous ovarian carcinoma. Proc
Natl Acad Sci USA. 112:E1272–E1277. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Buchanan EM, Weinstein LC and Hillson C:
Endometrial cancer. Am Fam Physician. 80:1075–1080. 2009.PubMed/NCBI
|
|
53
|
Setiawan VW, Yang HP, Pike MC, McCann SE,
Yu H, Xiang YB, Wolk A, Wentzensen N, Weiss NS, Webb PM, et al:
Type I and II endometrial cancers: Have they different risk
factors? J Clin Oncol. 31:2607–2618. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Matsumura N, Huang Z, Mori S, Baba T,
Fujii S, Konishi I, Iversen ES, Berchuck A and Murphy SK:
Epigenetic suppression of the TGF-beta pathway revealed by
transcriptome profiling in ovarian cancer. Genome Res. 21:74–82.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Talhouk A, McConechy MK, Leung S, Li-Chang
HH, Kwon JS, Melnyk N, Yang W, Senz J, Boyd N, Karnezis AN, et al:
A clinically applicable molecular-based classification for
endometrial cancers. Br J Cancer. 113:299–310. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Le Gallo M and Bell DW: The emerging
genomic landscape of endometrial cancer. Clin Chem. 60:98–110.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Garcia-Dios DA, Lambrechts D, Coenegrachts
L, Vandenput I, Capoen A, Webb PM, Ferguson K, Akslen LA, Claes B,
Vergote I, et al: Australian National Endometrial Cancer Study
Group: High-throughput interrogation of PIK3CA, PTEN, KRAS, FBXW7
and TP53 mutations in primary endometrial carcinoma. Gynecol Oncol.
128:327–334. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Hoang LN, McConechy MK, Köbel M, Han G,
Rouzbahman M, Davidson B, Irving J, Ali RH, Leung S, McAlpine JN,
et al: Histotype-genotype correlation in 36 high-grade endometrial
carcinomas. Am J Surg Pathol. 37:1421–1432. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Athanassiadou P, Athanassiades P, Grapsa
D, Gonidi M, Athanassiadou AM, Stamati PN and Patsouris E: The
prognostic value of PTEN, p53, and beta-catenin in endometrial
carcinoma: A prospective immunocytochemical study. Int J Gynecol
Cancer. 17:697–704. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Kuhn E, Wu RC, Guan B, Wu G, Zhang J, Wang
Y, Song L, Yuan X, Wei L, Roden RB, et al: Identification of
molecular pathway aberrations in uterine serous carcinoma by
genome-wide analyses. J Natl Cancer Inst. 104:1503–1513. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Zhao S, Choi M, Overton JD, Bellone S,
Roque DM, Cocco E, Guzzo F, English DP, Varughese J, Gasparrini S,
et al: Landscape of somatic single-nucleotide and copy-number
mutations in uterine serous carcinoma. Proc Natl Acad Sci USA.
110:2916–2921. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Le Gallo M, O'Hara AJ, Rudd ML, Urick ME,
Hansen NF, O'Neil NJ, Price JC, Zhang S, England BM, Godwin AK, et
al: Exome sequencing of serous endometrial tumors identifies
recurrent somatic mutations in chromatin-remodeling and ubiquitin
ligase complex genes. Nat Genet. 44:1310–1315. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Giannakis M, Hodis E, Mu X Jasmine,
Yamauchi M, Rosenbluh J, Cibulskis K, Saksena G, Lawrence MS, Qian
ZR, Nishihara R, et al: RNF43 is frequently mutated in colorectal
and endometrial cancers. Nat Genet. 46:1264–1266. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Markowska A, Pawałowska M, Lubin J and
Markowska J: Signalling pathways in endometrial cancer. Contemp
Oncol (Pozn). 18:143–148. 2014.PubMed/NCBI
|
|
65
|
Jo YS, Kim MS, Lee JH, Lee SH, An CH and
Yoo NJ: Frequent frameshift mutations in 2 mononucleotide repeats
of RNF43 gene and its regional heterogeneity in gastric and
colorectal cancers. Hum Pathol. 46:1640–1646. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Liu Y, Patel L, Mills GB, Lu KH, Sood AK,
Ding L, Kucherlapati R, Mardis ER, Levine DA, Shmulevich I, et al:
Clinical significance of CTNNB1 mutation and Wnt pathway activation
in endometrioid endometrial carcinoma. J Natl Cancer Inst.
106:dju2452014. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Carvajal-Carmona LG, O'Mara TA, Painter
JN, Lose FA, Dennis J, Michailidou K, Tyrer JP, Ahmed S, Ferguson
K, Healey CS, et al: Candidate locus analysis of the TERT-CLPTM1L
cancer risk region on chromosome 5p15 identifies multiple
independent variants associated with endometrial cancer risk. Hum
Genet. 134:231–245. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Fredriksson NJ, Ny L, Nilsson JA and
Larsson E: Systematic analysis of noncoding somatic mutations and
gene expression alterations across 14 tumor types. Nat Genet.
46:1258–1263. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Oshita T, Nagai N and Ohama K: Expression
of telomerase reverse transcriptase mRNA and its quantitative
analysis in human endometrial cancer. Int J Oncol. 17:1225–1230.
2000.PubMed/NCBI
|
|
70
|
Merritt MA and Cramer DW: Molecular
pathogenesis of endometrial and ovarian cancer. Cancer Biomark.
9:287–305. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Erickson BK, Kinde I, Dobbin ZC, Wang Y,
Martin JY, Alvarez RD, Conner MG, Huh WK, Roden RB, Kinzler KW, et
al: Detection of somatic TP53 mutations in tampons of patients with
high-grade serous ovarian cancer. Obstet Gynecol. 124:881–885.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
McConechy MK, Ding J, Cheang MC, Wiegand
KC, Senz J, Tone AA, Yang W, Prentice LM, Tse K, Zeng T, et al: Use
of mutation profiles to refine the classification of endometrial
carcinomas. J Pathol. 228:20–30. 2012.PubMed/NCBI
|
|
73
|
Parkinson DR, Johnson BE and Sledge GW:
Making personalized cancer medicine a reality: Challenges and
opportunities in the development of biomarkers and companion
diagnostics. Clin Cancer Res. 18:619–624. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Shih Ie M and Kurman RJ: Ovarian
tumorigenesis: A proposed model based on morphological and
molecular genetic analysis. Am J Pathol. 164:1511–1518. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Piek JM, van Diest PJ, Zweemer RP, et al:
Dysplastic changes in prophylactically removed Fallopian tubes of
women predisposed to developing ovarian cancer. J Pathol.
195:451–456. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Curtin N: PARP inhibitors for anticancer
therapy. Biochem Soc Trans. 42:82–88. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Cass I, Baldwin RL, Varkey T, Moslehi R,
Narod SA and Karlan BY: Improved survival in women with
BRCA-associated ovarian carcinoma. Cancer. 97:2187–2195. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Ashworth A: A synthetic lethal therapeutic
approach: Poly (ADP) ribose polymerase inhibitors for the treatment
of cancers deficient in DNA double-strand break repair. J Clin
Oncol. 126:3785–3790. 2008. View Article : Google Scholar
|
|
79
|
McCabe N, Turner NC, Lord CJ, Kluzek K,
Bialkowska A, Swift S, Giavara S, O'Connor MJ, Tutt AN, Zdzienicka
MZ, et al: Deficiency in the repair of DNA damage by homologous
recombination and sensitivity to poly(ADP-ribose) polymerase
inhibition. Cancer Res. 66:8109–8115. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Bryant HE, Schultz N, Thomas HD, Parker
KM, Flower D, Lopez E, Kyle S, Meuth M, Curtin NJ and Helleday T:
Specific killing of BRCA2-deficient tumours with inhibitors of
poly(ADP-ribose) polymerase. Nature. 434:913–917. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Gelmon KA, Tischkowitz M, Mackay H,
Swenerton K, Robidoux A, Tonkin K, Hirte H, Huntsman D, Clemons M,
Gilks B, et al: Olaparib in patients with recurrent high-grade
serous or poorly differentiated ovarian carcinoma or
triple-negative breast cancer: A phase 2, multicentre, open-label,
non-randomised study. Lancet Oncol. 12:852–861. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Oza AM, Cibula D, Benzaquen AO, Poole C,
Mathijssen RH, Sonke GS, Colombo N, Špaček J, Vuylsteke P, Hirte H,
et al: Olaparib combined with chemotherapy for recurrent
platinum-sensitive ovarian cancer: A randomised phase 2 trial.
Lancet Oncol. 16:87–97. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Lee JM, Hays JL, Annunziata CM, Noonan AM,
Minasian L, Zujewski JA, Yu M, Gordon N, Ji J, Sissung TM, et al:
Phase I/Ib study of olaparib and carboplatin in BRCA1 or BRCA2
mutation-associated breast or ovarian cancer with biomarker
analyses. J Natl Cancer Inst. 106:dju0892014. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Fingar DC, Richardson CJ, Tee AR, Cheatham
L, Tsou C and Blenis J: mTOR controls cell cycle progression
through its cell growth effectors S6K1 and 4E-BP1/eukaryotic
translation initiation factor 4E. Mol Cell Biol. 24:200–216. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Yuan TL and Cantley LC: PI3K pathway
alterations in cancer: Variations on a theme. Oncogene.
27:5497–5510. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Zoncu R, Efeyan A and Sabatini DM: mTOR:
From growth signal integration to cancer, diabetes and ageing. Nat
Rev Mol Cell Biol. 12:21–35. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Engelman JA: Targeting PI3K signalling in
cancer: Opportunities, challenges and limitations. Nat Rev Cancer.
9:550–562. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Motzer RJ, Escudier B, Oudard S, Hutson
TE, Porta C, Bracarda S, Grünwald V, Thompson JA, Figlin RA,
Hollaender N, et al: Phase 3 trial of everolimus for metastatic
renal cell carcinoma: Final results and analysis of prognostic
factors. Cancer. 116:4256–4265. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Husseinzadeh N and Husseinzadeh HD: mTOR
inhibitors and their clinical application in cervical, endometrial
and ovarian cancers: A critical review. Gynecol Oncol. 133:375–381.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Hirasawa T, Miyazawa M, Yasuda M, Shida M,
Ikeda M, Kajiwara H, Matsui N, Fujita M, Muramatsu T and Mikami M:
Alterations of hypoxia-induced factor signaling pathway due to
mammalian target of rapamycin (mTOR) suppression in ovarian clear
cell adenocarcinoma: In vivo and in vitro explorations for clinical
trial. Int J Gynecol Cancer. 23:1210–1218. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Köbel M, Huntsman D and Gilks CB: Critical
molecular abnormalities in high-grade serous carcinoma of the
ovary. Expert Rev Mol Med. 10:e222008. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Zoratto F, Rossi L, Giordani E, Strudel M,
Papa A and Tomao S: From conventional chemotherapy to targeted
therapy: Use of monoclonal antibodies (moAbs) in gastrointestinal
(GI) tumors. Tumour Biol. 35:8471–8482. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Kashiyama T, Oda K, Ikeda Y, Shiose Y,
Hirota Y, Inaba K, Makii C, Kurikawa R, Miyasaka A, Koso T, et al:
Antitumor activity and induction of TP53-dependent apoptosis toward
ovarian clear cell adenocarcinoma by the dual PI3K/mTOR inhibitor
DS-7423. PLoS One. 9:e872202014. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Shoji K, Oda K, Kashiyama T, Ikeda Y,
Nakagawa S, Sone K, Miyamoto Y, Hiraike H, Tanikawa M, Miyasaka A,
et al: Genotype-dependent efficacy of a dual PI3K/mTOR inhibitor,
NVP-BEZ235 and an mTOR inhibitor, RAD001, in endometrial
carcinomas. PLoS One. 7:e374312012. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Gershenson DM, Sun CC, Bodurka D, Coleman
RL, Lu KH, Sood AK, Deavers M, Malpica AL and Kavanagh JJ:
Recurrent low-grade serous ovarian carcinoma is relatively
chemoresistant. Gynecol Oncol. 114:48–52. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Gershenson DM, Sun CC, Lu KH, Coleman RL,
Sood AK, Malpica A, Deavers MT, Silva EG and Bodurka DC: Clinical
behavior of stage II–IV low-grade serous carcinoma of the ovary.
Obstet Gynecol. 108:361–368. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Miller CR, Oliver KE and Farley JH: MEK1/2
inhibitors in the treatment of gynecologic malignancies. Gynecol
Oncol. 133:128–137. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Farley J, Brady WE, Vathipadiekal V,
Lankes HA, Coleman R, Morgan MA, Mannel R, Yamada SD, Mutch D,
Rodgers WH, et al: Selumetinib in women with recurrent low-grade
serous carcinoma of the ovary or peritoneum: An open-label,
single-arm, phase 2 study. Lancet Oncol. 14:134–140. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Kuo KT, Guan B, Feng Y, Mao TL, Chen X,
Jinawath N, Wang Y, Kurman RJ, Shih Ie M and Wang TL: Analysis of
DNA copy number alterations in ovarian serous tumors identifies new
molecular genetic changes inlow-grade and high-grade carcinomas.
Cancer Res. 69:4036–4042. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Hamilton MP, Rajapakshe K, Hartig SM, Reva
B, McLellan MD, Kandoth C, Ding L, Zack TI, Gunaratne PH, Wheeler
DA, et al: Identification of a pan-cancer oncogenic microRNA
superfamily anchored by a central core seed motif. Nat Commun.
4:27302013. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Jacobsen A, Silber J, Harinath G, Huse JT,
Schultz N and Sander C: Analysis of microRNA-target interactions
across diverse cancer types. Nat Struct Mol Biol. 20:1325–1332.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Zovoilis A, Mungall AJ, Moore R, Varhol R,
Chu A, Wong T, Marra M and Jones SJ: The expression level of small
non-coding RNAs derived from the first exon of protein-coding genes
is predictive of cancer status. EMBO Rep. 15:402–410. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Mukherji S, Ebert MS, Zheng GX, Tsang JS,
Sharp PA and van Oudenaarden A: MicroRNAs can generate thresholds
in target gene expression. Nat Genet. 43:854–859. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Boren T, Xiong Y, Hakam A, Wenham R, Apte
S, Wei Z, Kamath S, Chen DT, Dressman H and Lancaster JM: MicroRNAs
and their target messenger RNAs associated with endometrial
carcinogenesis. Gynecol Oncol. 110:206–215. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Wu W, Lin Z, Zhuang Z and Liang X:
Expression profile of mammalian microRNAs in endometrioid
adenocarcinoma. Eur J Cancer Prev. 18:50–55. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Mauel S, Kruse B, Etschmann B, von der
Schulenburg AG, Schaerig M, Stövesand K, Wilcken B and Sterner-Kock
A: Latent transforming growth factor binding protein 4 (LTBP-4) is
downregulated in human mammary adenocarcinomas in vitro and in
vivo. APMIS. 115:687–700. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Burns MB, Temiz NA and Harris RS: Evidence
for APOBEC3B mutagenesis in multiple human cancers. Nat Genet.
45:977–983. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Roberts SA, Lawrence MS, Klimczak LJ,
Grimm SA, Fargo D, Stojanov P, Kiezun A, Kryukov GV, Carter SL,
Saksena G, et al: An APOBEC cytidine deaminase mutagenesis pattern
is widespread in human cancers. Nat Genet. 45:970–976. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Genome Atlas Research Network Cancer.
Weinstein JN, Collisson EA, Mills GB, Shaw KR, Ozenberger BA,
Ellrott K, Shmulevich I, Sander C and Stuart JM: The cancer genome
atlas pan-cancer analysis project. Nat Genet. 45:1113–1120. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Ren X, McHale CM, Skibola CF, Smith AH,
Smith MT and Zhang L: An emerging role for epigenetic dysregulation
in arsenic toxicity and carcinogenesis. Environ Health Perspect.
119:11–19. 2011. View Article : Google Scholar : PubMed/NCBI
|