|
1
|
Ross JS, Slodkowska EA, Symmans WF,
Pusztai L, Ravdin PM and Hortobágyi GN: The HER-2 receptor and
breast cancer: ten years of targeted anti-HER-2 therapy and
personalized medicine. Oncologist. 14:320–368. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Slamon DJ, Clark GM, Wong SG, Levin WJ,
Ullrich A and McGuire WL: Human breast cancer: correlation of
relapse and survival with amplification of the HER-2/neu oncogene.
Science. 235:177–182. 1987. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Slamon DJ, Leyland-Jones B, Shak S, Fuchs
H, Paton V, Bajamonde A, Fleming T, Eiermann W, Wolter J, Pegram M,
et al: Use of chemotherapy plus a monoclonal antibody against HER2
for metastatic breast cancer that overexpresses HER2. N Engl J Med.
344:783–792. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Perez EA, Romond EH, Suman VJ, Jeong JH,
Sledge G, Geyer CE Jr, Martino S, Rastogi P, Gralow J, Swain SM, et
al: Trastuzumab plus adjuvant chemotherapy for human epidermal
growth factor receptor 2-positive breast cancer: planned joint
analysis of overall survival from NSABP B-31 and NCCTG N9831. J
Clin Oncol. 32:3744–3752. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Rastogi P, Anderson SJ, Bear HD, Geyer CE,
Kahlenberg MS, Robidoux A, Margolese RG, Hoehn JL, Vogel VG, Dakhil
SR, et al: Preoperative chemotherapy: updates of national surgical
adjuvant breast and bowel project protocols B-18 and B-27. J Clin
Oncol. 26:778–785. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kong X, Moran MS, Zhang N, Haffty B and
Yang Q: Meta-analysis confirms achieving pathological complete
response after neoadjuvant chemotherapy predicts favourable
prognosis for breast cancer patients. Eur J Cancer. 47:2084–2090.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Buzdar AU: Significantly higher pathologic
complete remission rate after neoadjuvant therapy with trastuzumab,
paclitaxel, and epirubicin chemotherapy: results of a randomized
trial in human epidermal growth factor receptor 2-positive operable
breast cancer. J Clin Oncol. 23:3676–3685. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Gianni L, Eiermann W, Semiglazov V,
Manikhas A, Lluch A, Tjulandin S, Zambetti M, Vazquez F, Byakhow M,
Lichinitser M, et al: Neoadjuvant chemotherapy with trastuzumab
followed by adjuvant trastuzumab versus neoadjuvant chemotherapy
alone, in patients with HER2-positive locally advanced breast
cancer (the NOAH trial): a randomised controlled superiority trial
with a parallel HER2-negative cohort. Lancet. 375:377–384. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Goldhirsch A, Winer EP, Coates AS, Gelber
RD, Piccart-Gebhart M, Thürlimann B and Senn HJ: Panel members:
Personalizing the treatment of women with early breast cancer:
highlights of the St Gallen international expert consensus on the
primary therapy of early breast cancer, 2013. Ann Oncol.
24:2206–2223. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Cortazar P, Zhang L, Untch M, Mehta K,
Costantino JP, Wolmark N, Bonnefoi H, Cameron D, Gianni L,
Valagussa P, et al: Pathological complete response and long-term
clinical benefit in breast cancer: the CTNeoBC pooled analysis.
Lancet. 384:164–172. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Mulrane L, McGee SF, Gallagher WM and
O'Connor DP: miRNA dysregulation in breast cancer. Cancer Res.
73:6554–6562. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wang SE and Lin RJ: MicroRNA and
HER2-overexpressing cancer. Microrna. 2:137–147. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Serpico D, Molino L and Di Cosimo S:
microRNAs in breast cancer development and treatment. Cancer Treat
Rev. 40:595–604. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Blenkiron C, Goldstein LD, Thorne NP,
Spiteri I, Chin SF, Dunning MJ, Barbosa-Morais NL, Teschendorff AE,
Green AR, Ellis IO, et al: MicroRNA expression profiling of human
breast cancer identifies new markers of tumor subtype. Genome Biol.
8:R2142007. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Dai X, Chen A and Bai Z: Integrative
investigation on breast cancer in ER, PR and HER2-defined subgroups
using mRNA and miRNA expression profiling. Sci Rep. 4:65662014.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wolff AC, Hammond MEH, Schwartz JN,
Hagerty KL, Allred DC, Cote RJ, Dowsett M, Fitzgibbons PL, Hanna
WM, Langer A, et al: American Society of Clinical Oncology/College
of American Pathologists guideline recommendations for human
epidermal growth factor receptor 2 testing in breast cancer. J Clin
Oncol. 25:118–145. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ni X, Xia T, Zhao Y, Zhou W, Wu N, Liu X,
Ding Q, Zha X, Sha J and Wang S: Downregulation of miR-106b induced
breast cancer cell invasion and motility in association with
overexpression of matrix metalloproteinase 2. Cancer Sci.
105:18–25. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Isobe T, Hisamori S, Hogan DJ, Zabala M,
Hendrickson DG, Dalerba P, Cai S, Scheeren F, Kuo AH, Sikandar SS,
et al: miR-142 regulates the tumorigenicity of human breast cancer
stem cells through the canonical WNT signaling pathway. Elife.
3:2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Huang S, Chen Y, Wu W, Ouyang N, Chen J,
Li H, Liu X, Su F, Lin L and Yao Y: miR-150 promotes human breast
cancer growth and malignant behavior by targeting the pro-apoptotic
purinergic P2X7 receptor. PLoS One. 8:e807072013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lowery AJ, Miller N, Devaney A, McNeill
RE, Davoren PA, Lemetre C, Benes V, Schmidt S, Blake J, Ball G and
Kerin MJ: MicroRNA signatures predict oestrogen receptor,
progesterone receptor and HER2/neu receptor status in breast
cancer. Breast Cancer Res. 11:R272009. View
Article : Google Scholar : PubMed/NCBI
|
|
21
|
Madhavan D, Zucknick M, Wallwiener M, Cuk
K, Modugno C, Scharpff M, Schott S, Heil J, Turchinovich A, Yang R,
et al: Circulating miRNAs as surrogate markers for circulating
tumor cells and prognostic markers in metastatic breast cancer.
Clin Cancer Res. 18:5972–5982. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Li M, Ma X, Li M, Zhang B, Huang J, Liu L
and Wei Y: Prognostic role of microRNA-210 in various carcinomas: a
systematic review and meta-analysis. Dis Markers. 2014:1061972014.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
He X, Xiao X, Dong L, Wan N, Zhou Z, Deng
H and Zhang X: MiR-218 regulates cisplatin chemosensitivity in
breast cancer by targeting BRCA1. Tumour Biol. 36:2065–2075. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Lv J, Xia K, Xu P, Sun E, Ma J, Gao S,
Zhou Q, Zhang M, Wang F, Chen F, et al: miRNA expression patterns
in chemoresistant breast cancer tissues. Biomed Pharmacother.
68:935–942. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yamamoto Y, Yoshioka Y, Minoura K,
Takahashi RU, Takeshita F, Taya T, Horii R, Fukuoka Y, Kato T,
Kosaka N and Ochiya T: An integrative genomic analysis revealed the
relevance of microRNA and gene expression for drug-resistance in
human breast cancer cells. Mol Cancer. 10:1352011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Shen J, Hu Q, Schrauder M, Yan L, Wang D,
Medico L, Guo Y, Yao S, Zhu Q, Liu B, et al: Circulating miR-148b
and miR-133a as biomarkers for breast cancer detection. Oncotarget.
5:5284–5294. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Pinto R, De Summa S, Danza K, Popescu O,
Paradiso A, Micale L, Merla G, Palumbo O, Carella M and Tommasi S:
MicroRNA expression profiling in male and female familial breast
cancer. Br J Cancer. 111:2361–2368. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Shi W, Gerster K, Alajez NM, Tsang J,
Waldron L, Pintilie M, Hui AB, Sykes J, P'ng C, Miller N, et al:
MicroRNA-301 mediates proliferation and invasion in human breast
cancer. Cancer Res. 71:2926–2937. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kim NH, Kim HS, Li XY, Lee I, Choi HS,
Kang SE, Cha SY, Ryu JK, Yoon D, Fearon ER, et al: A p53/miRNA-34
axis regulates Snail1-dependent cancer cell epithelial-mesenchymal
transition. J Cell Biol. 195:417–433. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Cuk K, Zucknick M, Heil J, Madhavan D,
Schott S, Turchinovich A, Arlt D, Rath M, Sohn C, Benner A, et al:
Circulating microRNAs in plasma as early detection markers for
breast cancer. Int J Cancer. 132:1602–1612. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Kim SJ, Shin JY, Lee KD, Bae YK, Sung KW,
Nam SJ and Chun KH: MicroRNA let-7a suppresses breast cancer cell
migration and invasion through downregulation of C-C chemokine
receptor type 7. Breast Cancer Res. 14:R142012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Pérez-Rivas LG, Jerez JM, Carmona R, de
Luque V, Vicioso L, Claros MG, Viguera E, Pajares B, Sánchez A,
Ribelles N, et al: A microRNA signature associated with early
recurrence in breast cancer. PLoS One. 9:e918842014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Perou CM, Sørlie T, Eisen MB, van de Rijn
M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA,
et al: Molecular portraits of human breast tumours. Nature.
406:747–752. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Jung EJ, Santarpia L, Kim J, Esteva FJ,
Moretti E, Buzdar AU, Di Leo A, Le XF, Bast RC Jr, Park ST, et al:
Plasma microRNA 210 levels correlate with sensitivity to
trastuzumab and tumor presence in breast cancer patients. Cancer.
118:2603–2614. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Müller V, Gade S, Steinbach B, Loibl S,
von Minckwitz G, Untch M, Schwedler K, Lübbe K, Schem C, Fasching
PA, et al: Changes in serum levels of miR-21, miR-210 and miR-373
in HER2-positive breast cancer patients undergoing neoadjuvant
therapy: a translational research project within the Geparquinto
trial. Breast Cancer Res Treat. 147:61–68. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Camps C, Buffa FM, Colella S, Moore J,
Sotiriou C, Sheldon H, Harris AL, Gleadle JM and Ragoussis J:
hsa-miR-210 is induced by hypoxia and is an independent prognostic
factor in breast cancer. Clin Cancer Res. 14:1340–1348. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
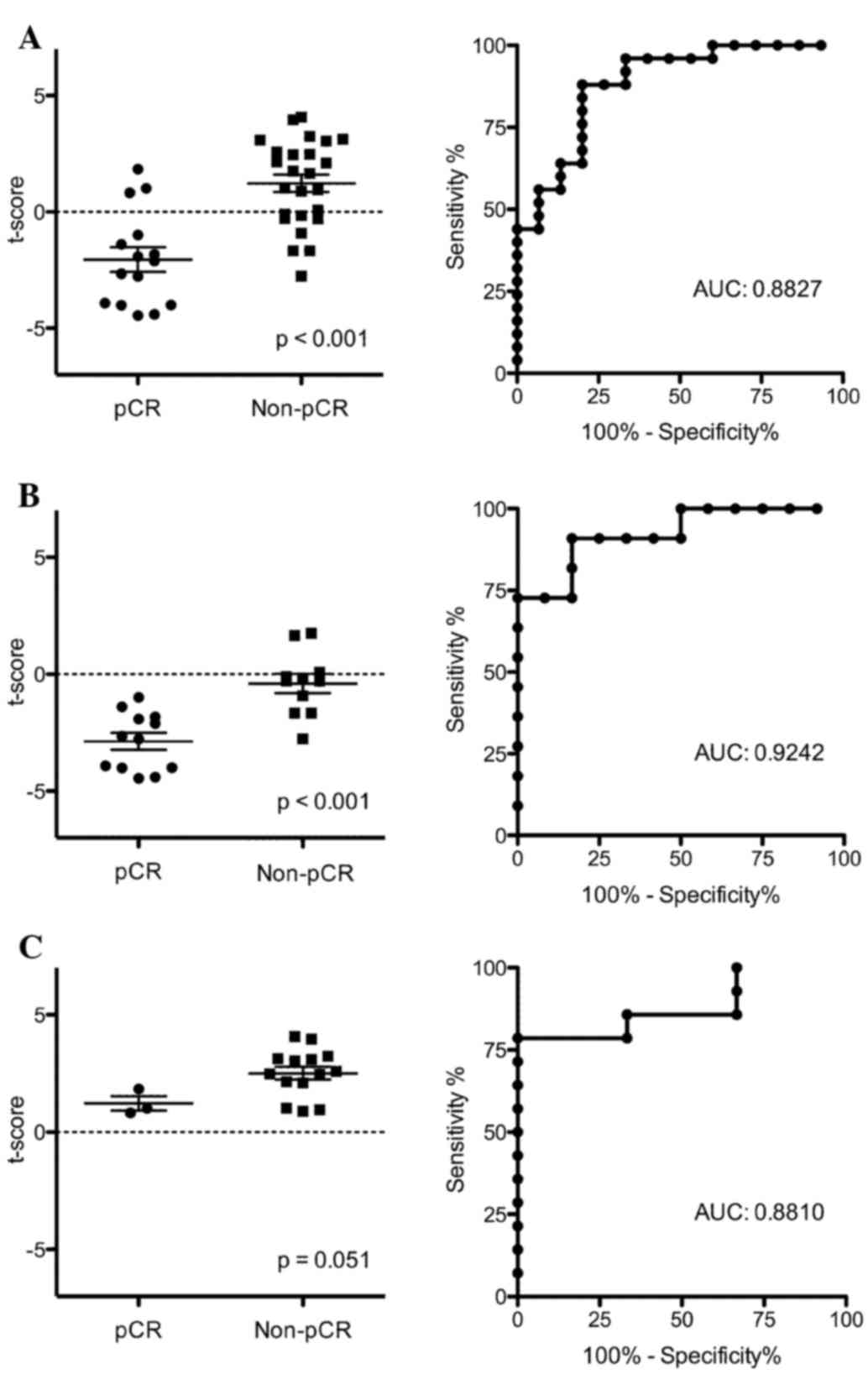
Kolacinska A, Morawiec J, Fendler W,
Malachowska B, Morawiec Z, Szemraj J, Pawlowska Z, Chowdhury D,
Choi YE, Kubiak R, et al: Association of microRNAs and pathologic
response to preoperative chemotherapy in triple negative breast
cancer: preliminary report. Mol Biol Rep. 41:2851–2857. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Sotiriou C and Pusztai L: Gene-expression
signatures in breast cancer. N Engl J Med. 360:790–800. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Mitchell PS, Parkin RK, Kroh EM, Fritz BR,
Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O'Briant
KC, Allen A, et al: Circulating microRNAs as stable blood-based
markers for cancer detection. Proc Natl Acad Sci USA.
105:10513–10518. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K,
Guo J, Zhang Y, Chen J, Guo X, et al: Characterization of microRNAs
in serum: a novel class of biomarkers for diagnosis of cancer and
other diseases. Cell Res. 18:997–1006. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Pritchard CC, Cheng HH and Tewari M:
MicroRNA profiling: approaches and considerations. Nat Rev Genet.
13:358–369. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
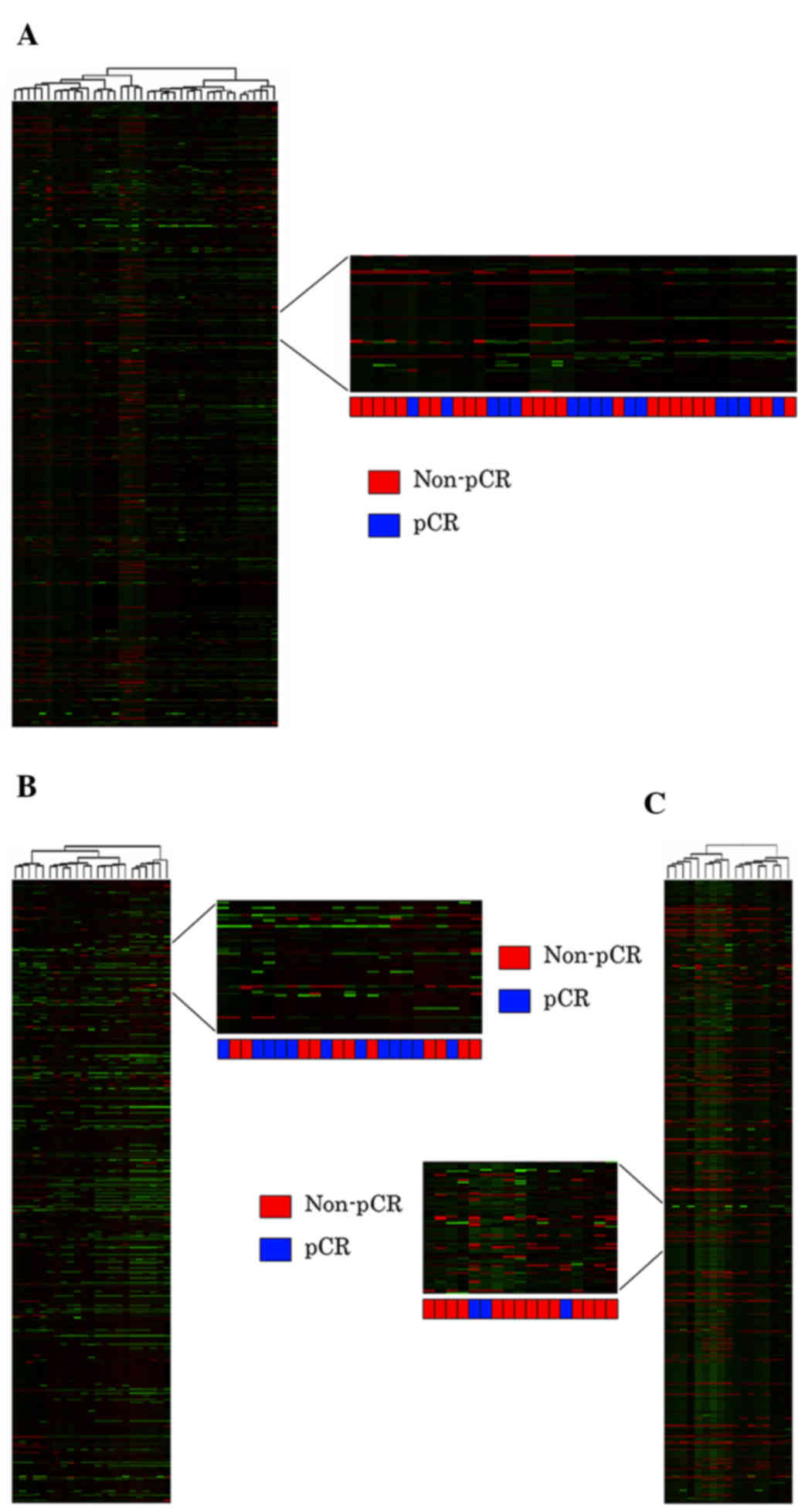
Hui AB, Shi W, Boutros PC, Miller N,
Pintilie M, Fyles T, McCready D, Wong D, Gerster K, Waldron L, et
al: Robust global micro-RNA profiling with formalin-fixed
paraffin-embedded breast cancer tissues. Lab Invest. 89:597–606.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Xi Y, Nakajima G, Gavin E, Morris CG, Kudo
K, Hayashi K and Ju J: Systematic analysis of microRNA expression
of RNA extracted from fresh frozen and formalin-fixed
paraffin-embedded samples. RNA. 13:1668–1674. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
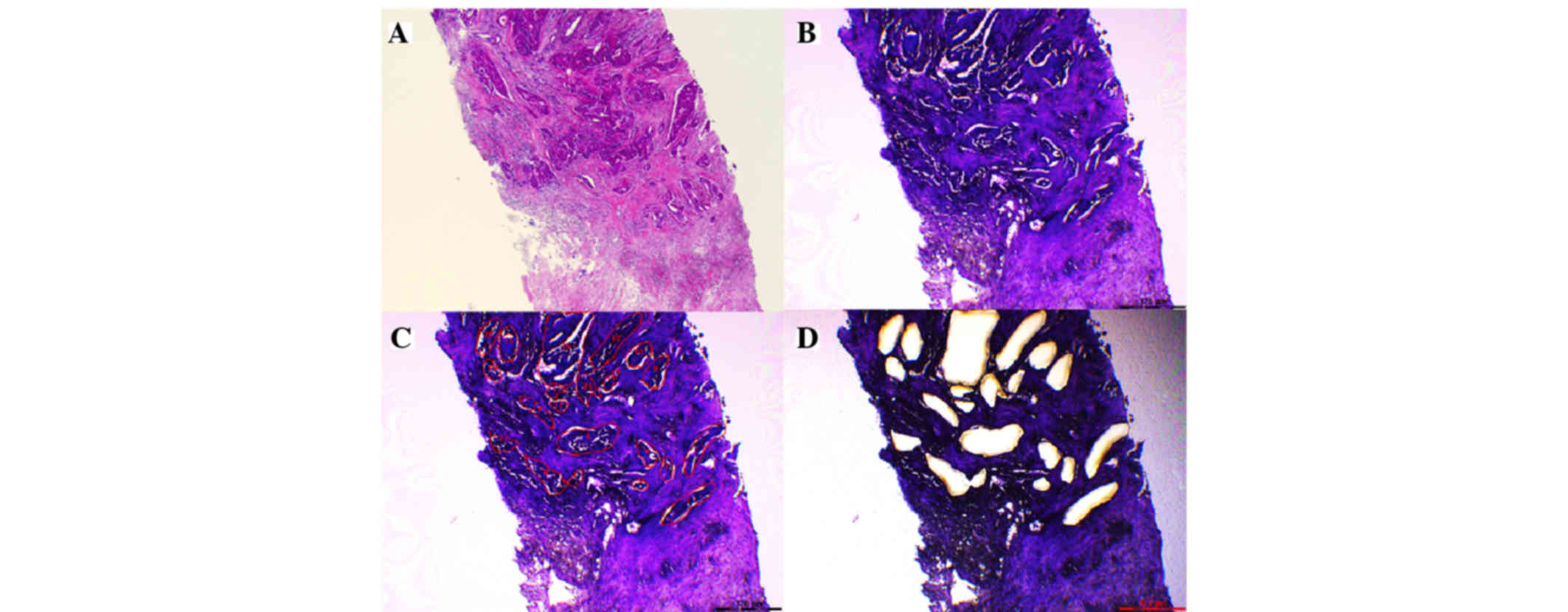
Legres LG, Janin A, Masselon C and
Bertheau P: Beyond laser microdissection technology: follow the
yellow brick road for cancer research. Am J Cancer Res. 4:1–28.
2014.PubMed/NCBI
|
|
45
|
Lassmann S, Kreutz C, Schoepflin A, Hopt
U, Timmer J and Werner M: A novel approach for reliable microarray
analysis of microdissected tumor cells from formalin-fixed and
paraffin-embedded colorectal cancer resection specimens. J Mol Med
(Berl). 87:211–224. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Meyer SU, Pfaffl MW and Ulbrich SE:
Normalization strategies for microRNA profiling experiments: a
‘normal’ way to a hidden layer of complexity? Biotechnol Lett.
32:1777–1788. 2010. View Article : Google Scholar : PubMed/NCBI
|