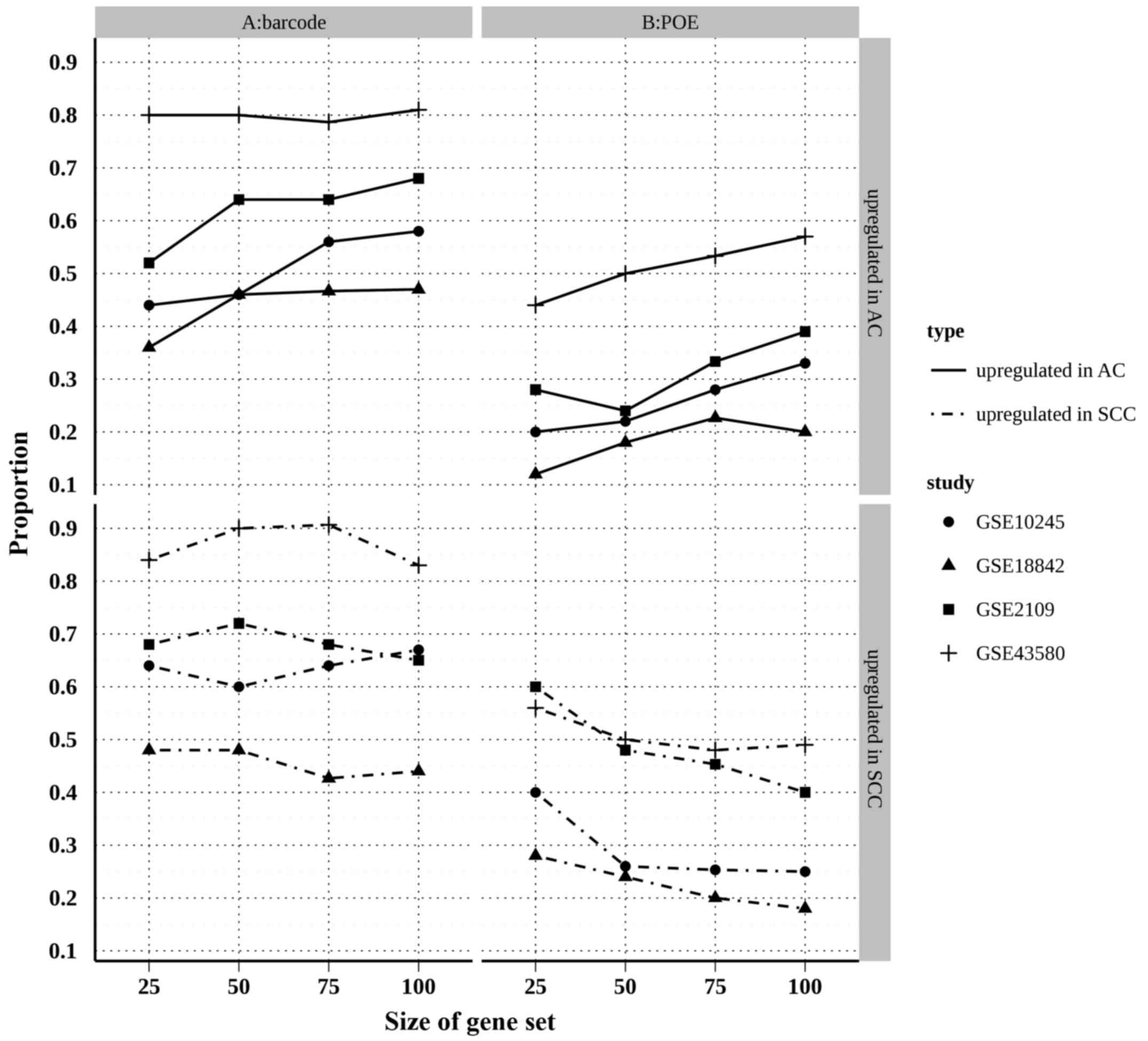
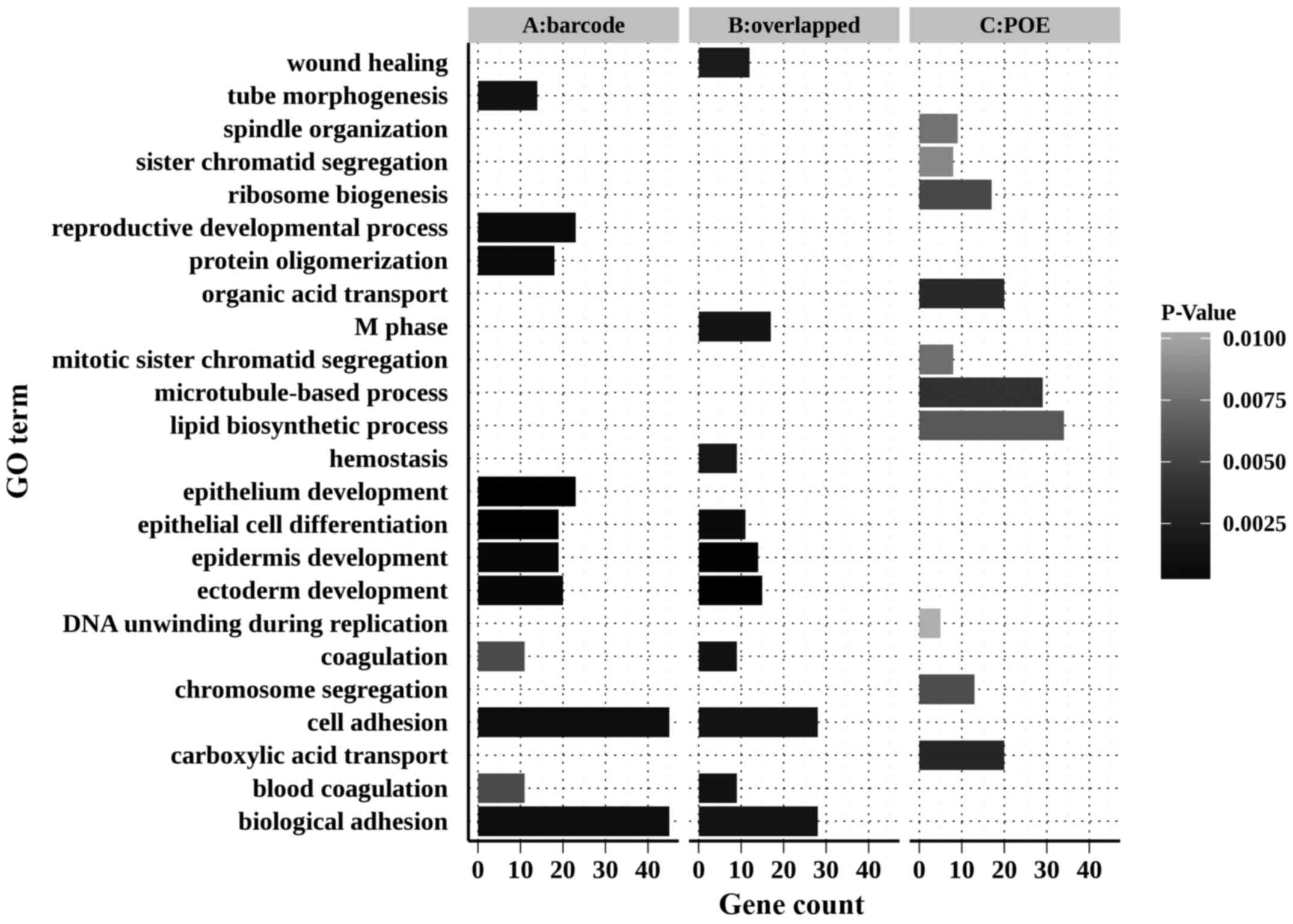
|
1
|
Kikuchi T, Daigo Y, Katagiri T, Tsunoda T,
Okada K, Kakiuchi S, Zembutsu H, Furukawa Y, Kawamura M, Kobayashi
K, et al: Expression profiles of non-small cell lung cancers on
cDNA microarrays: Identification of genes for prediction of
lymph-node metastasis and sensitivity to anti-cancer drugs.
Oncogene. 22:2192–2205. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Lemjabbar-Alaoui H, Hassan OU, Yang YW and
Buchanan P: Lung cancer: Biology and treatment options. Biochim
Biophys Acta. 1856:189–210. 2015.PubMed/NCBI
|
|
3
|
Liu J, Yang XY and Shi WJ: Identifying
differentially expressed genes and pathways in two types of
non-small cell lung cancer: Adenocarcinoma and squamous cell
carcinoma. Genet Mol Res. 13:95–102. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kuner R, Muley T, Meister M, Ruschhaupt M,
Buness A, Xu EC, Schnabel P, Warth A, Poustka A, Sültmann H and
Hoffmann H: Global gene expression analysis reveals specific
patterns of cell junctions in non-small cell lung cancer subtypes.
Lung Cancer. 63:32–38. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Li L, Zhu J, Guo SX and Deng Y: Bicluster
and regulatory network analysis of differentially expressed genes
in adenocarcinoma and squamous cell carcinoma. Genet Mol Res.
12:1710–1719. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Li J, Li D, Wei X and Su Y: In silico
comparative genomic analysis of two non-small cell lung cancer
subtypes and their potentials for cancer classification. Cancer
Genomics Proteomics. 11:303–310. 2014.PubMed/NCBI
|
|
7
|
Siddiqui AS, Delaney AD, Schnerch A,
Griffith OL, Jones SJ and Marra MA: Sequence biases in large scale
gene expression profiling data. Nucleic Acids Res. 34:e832006.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Choi JK, Yu U, Kim S and Yoo OJ: Combining
multiple microarray studies and modeling interstudy variation.
Bioinformatics. 19:(Suppl 1). i84–i90. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Rhodes DR, Barrette TR, Rubin MA, Ghosh D
and Chinnaiyan AM: Meta-analysis of microarrays: Interstudy
validation of gene expression profiles reveals pathway
dysregulation in prostate cancer. Cancer Res. 62:4427–4433.
2002.PubMed/NCBI
|
|
10
|
Choi H, Shen R, Chinnaiyan AM and Ghosh D:
A latent variable approach for meta-analysis of gene expression
data from multiple microarray experiments. BMC Bioinformatics.
8:3642007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zilliox MJ and Irizarry RA: A gene
expression bar code for microarray data. Nat Methods. 4:911–913.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Amelung JT, Bührens R, Beshay M and
Reymond MA: Key genes in lung cancer translational research: A
meta-analysis. Pathobiology. 77:53–63. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kim B, Lee HJ, Choi HY, Shin Y, Nam S, Seo
G, Son DS, Jo J, Kim J, Lee J, et al: Clinical validity of the lung
cancer biomarkers identified by bioinformatics analysis of public
expression data. Cancer Res. 67:7431–7438. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Tian ZQ, Li ZH, Wen SW, Zhang YF, Li Y,
Cheng JG and Wang GY: Identification of commonly dysregulated genes
in non-small-cell lung cancer by integrated analysis of microarray
data and qRT-PCR validation. Lung. 193:583–592. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Tan X and Chen M: MYLK and MYL9 expression
in non-small cell lung cancer identified by bioinformatics analysis
of public expression data. Tumour Biol. 35:12189–12200. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Huang CH, Chang PM, Lin YJ, Wang CH, Huang
CY and Ng KL: Drug repositioning discovery for early- and
late-stage non-small-cell lung cancer. Biomed Res Int.
2014:1938172014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
McCall MN, Bolstad BM and Irizarry RA:
Frozen robust multiarray analysis (fRMA). Biostatistics.
11:242–253. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Lee JK, Bussey KJ, Gwadry FG, Reinhold W,
Riddick G, Pelletier SL, Nishizuka S, Szakacs G, Annereau JP,
Shankavaram U, et al: Comparing cDNA and oligonucleotide array
data: Concordance of gene expression across platforms for the
NCI-60 cancer cells. Genome Biol. 4:R822003. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Parmigiani G, Garrett-Mayer ES, Anbazhagan
R and Gabrielson E: A cross-study comparison of gene expression
studies for the molecular classification of lung cancer. Clin
Cancer Res. 10:2922–2927. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Borenstein M, Hedges LV, Higgins JPT and
Rothstein HR: Introduction to Meta-Analysis. 1st. John Wiley and
Sons, Ltd.; Chichester, UK: 2009, View Article : Google Scholar
|
|
21
|
McCall MN, Uppal K, Jaffee HA, Zilliox MJ
and Irizarry RA: The gene expression barcode: Leveraging public
data repositories to begin cataloging the human and murine
transcriptomes. Nucleic Acids Res. 39:D1011–D1015. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Cochran WG: The comparison of percentages
in matched samples. Biometrika. 37:256–266. 1950. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Stevens JR and Doerge RW: Combining
Affymetrix microarray results. BMC Bioinformatics. 6:572005.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Conlon EM, Song JJ and Liu A: Bayesian
meta-analysis models for microarray data: A comparative study. BMC
Bioinformatics. 8:802007. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Tusher VG, Tibshirani R and Chu G:
Significance analysis of microarrays applied to the ionizing
radiation response. Proc Natl Acad Sci USA. 98:5116–5121. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Hong F and Breitling R: A comparison of
meta-analysis methods for detecting differentially expressed genes
in microarray experiments. Bioinformatics. 24:374–382. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Irizarry RA, Warren D, Spencer F, Kim IF,
Biswal S, Frank BC, Gabrielson E, Garcia JG, Geoghegan J, Germino
G, et al: Multiple-laboratory comparison of microarray platforms.
Nat Methods. 2:345–350. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
da Huang W, Sherman BT and Lempicki RA:
Bioinformatics enrichment tools: Paths toward the comprehensive
functional analysis of large gene lists. Nucleic Acids Res.
37:1–13. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
da Huang W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Kwei KA, Kim YH, Girard L, Kao J,
Pacyna-Gengelbach M, Salari K, Lee J, Choi YL, Sato M, Wang P, et
al: Genomic profiling identifies TITF1 as a lineage-specific
oncogene amplified in lung cancer. Oncogene. 27:3635–3640. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Massion PP, Taflan PM, Rahman Jamshedur
SM, Yildiz P, Shyr Y, Edgerton ME, Westfall MD, Roberts JR,
Pietenpol JA, Carbone DP and Gonzalez AL: Significance of p63
amplification and overexpression in lung cancer development and
prognosis. Cancer Res. 63:7113–7121. 2003.PubMed/NCBI
|
|
32
|
Zhou ZY, Yang GY, Zhou J and Yu MH:
Significance of TRIM29 and β-catenin expression in non-small-cell
lung cancer. J Chin Med Assoc. 75:269–274. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Angulo B, Suarez-Gauthier A, Lopez-Rios F,
Medina PP, Conde E, Tang M, Soler G, Lopez-Encuentra A, Cigudosa JC
and Sanchez-Cespedes M: Expression signatures in lung cancer reveal
a profile for EGFR-mutant tumours and identify selective PIK3CA
overexpression by gene amplification. J Pathol. 214:347–356. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Li Y, Wei S, Wang J, Hong L, Cui L and
Wang C: Analysis of the factors associated with abnormal
coagulation and prognosis in patients with non-small cell lung
cancer. Zhongguo Fei Ai Za Zhi. 17:789–796. 2014.(In Chinese).
PubMed/NCBI
|
|
35
|
Kogan EA, Ugriumov DA and Jaques G:
Morphologic and molecular-genetic characteristics of keratinization
and apoptosis in squamous cell lung carcinoma. Arkh Patol.
62:16–20. 2000.PubMed/NCBI
|
|
36
|
Merikallio H, Pääkkö P, Harju T and Soini
Y: Claudins 10 and 18 are predominantly expressed in lung
adenocarcinomas and in tumors of nonsmokers. Int J Clin Exp Pathol.
4:667–673. 2011.PubMed/NCBI
|
|
37
|
Kurotani R, Tomita T, Yang Q, Carlson BA,
Chen C and Kimura S: Role of secretoglobin 3A2 in lung development.
Am J Respir Crit Care Med. 178:389–398. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Davidson B, Stavnes HT, Risberg B, Nesland
JM, Wohlschlaeger J, Yang Y, Shih IeM and Wang TL: Gene expression
signatures differentiate adenocarcinoma of lung and breast origin
in effusions. Hum Pathol. 43:684–694. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Vischioni B, Oudejans JJ, Vos W, Rodriguez
JA and Giaccone G: Frequent overexpression of aurora B kinase, a
novel drug target, in non-small cell lung carcinoma patients. Mol
Cancer Ther. 5:2905–2913. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Lane EB and McLean WH: Keratins and skin
disorders. J Pathol. 204:355–366. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Landi MT, Chatterjee N, Yu K, Goldin LR,
Goldstein AM, Rotunno M, Mirabello L, Jacobs K, Wheeler W, Yeager
M, et al: A genome-wide association study of lung cancer identifies
a region of chromosome 5p15 associated with risk for
adenocarcinoma. Am J Hum Genet. 85:679–691. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Hsiung CA, Lan Q, Hong YC, Chen CJ,
Hosgood HD, Chang IS, Chatterjee N, Brennan P, Wu C, Zheng W, et
al: The 5p15.33 locus is associated with risk of lung
adenocarcinoma in never-smoking females in Asia. PLoS Genet. 6:pii:
e1001051. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Ni Z, Tao K, Chen G, Chen Q, Tang J, Luo
X, Yin P, Tang J and Wang X: CLPTM1L is overexpressed in lung
cancer and associated with apoptosis. PLoS One. 7:e525982012.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Dong J, Jin G, Wu C, Guo H, Zhou B, Lv J,
Lu D, Shi Y, Shu Y, Xu L, et al: Genome-wide association study
identifies a novel susceptibility locus at 12q23.1 for lung
squamous cell carcinoma in han Chinese. PLoS Genet. 9:e10031902013.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Modica S, Murzilli S, Salvatore L, Schmidt
DR and Moschetta A: Nuclear bile acid receptor FXR protects against
intestinal tumorigenesis. Cancer Res. 68:9589–9594. 2008.
View Article : Google Scholar : PubMed/NCBI
|