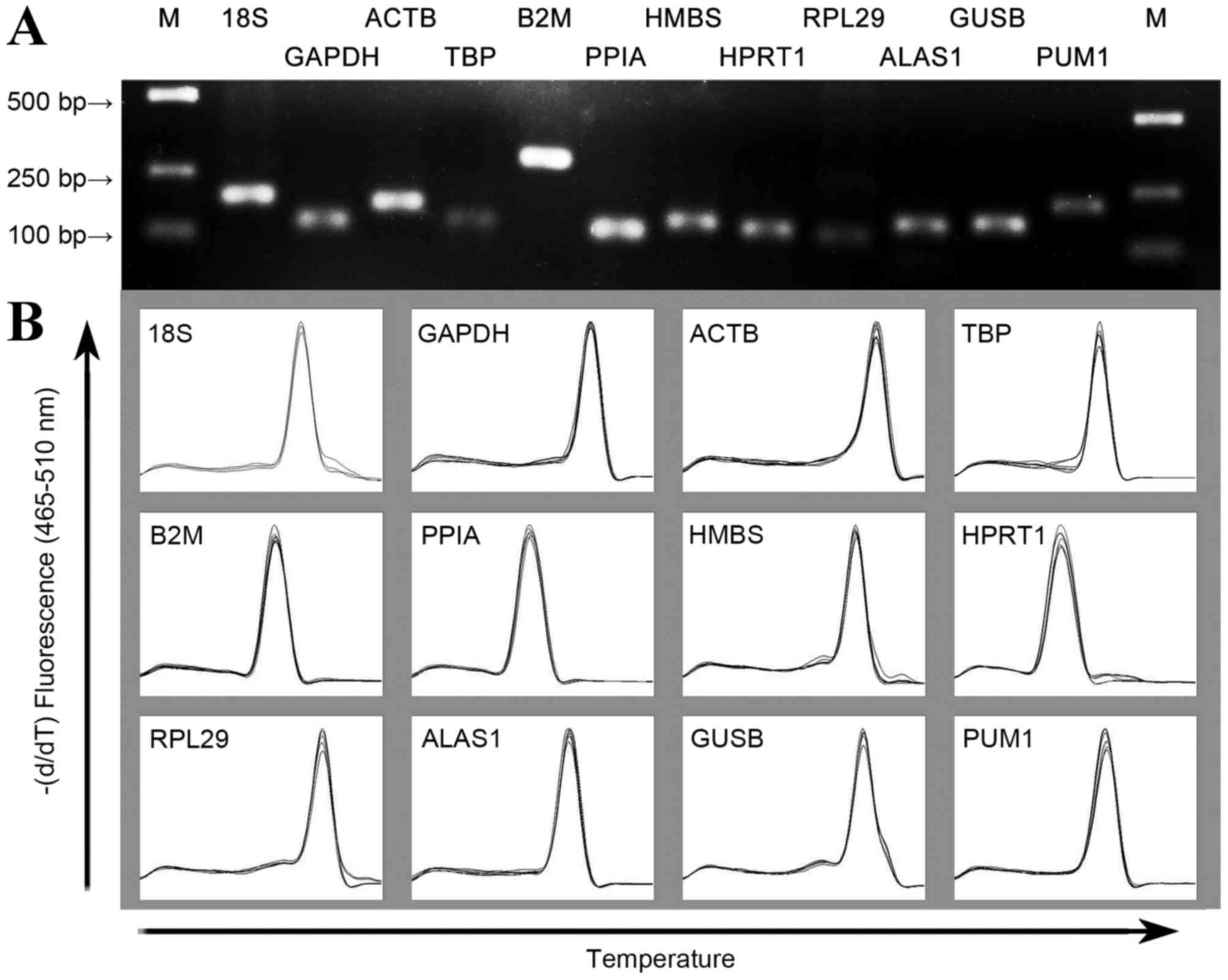
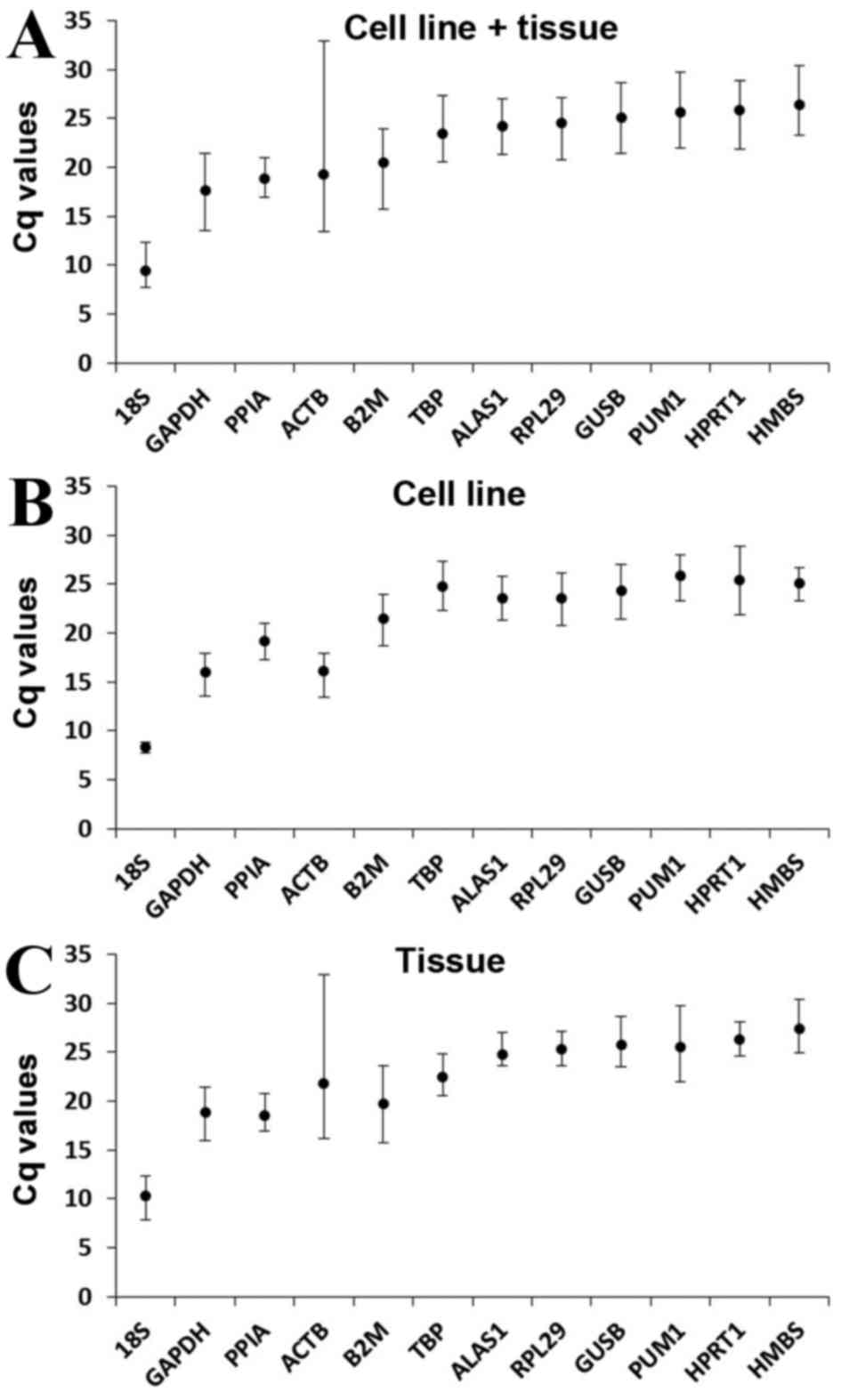
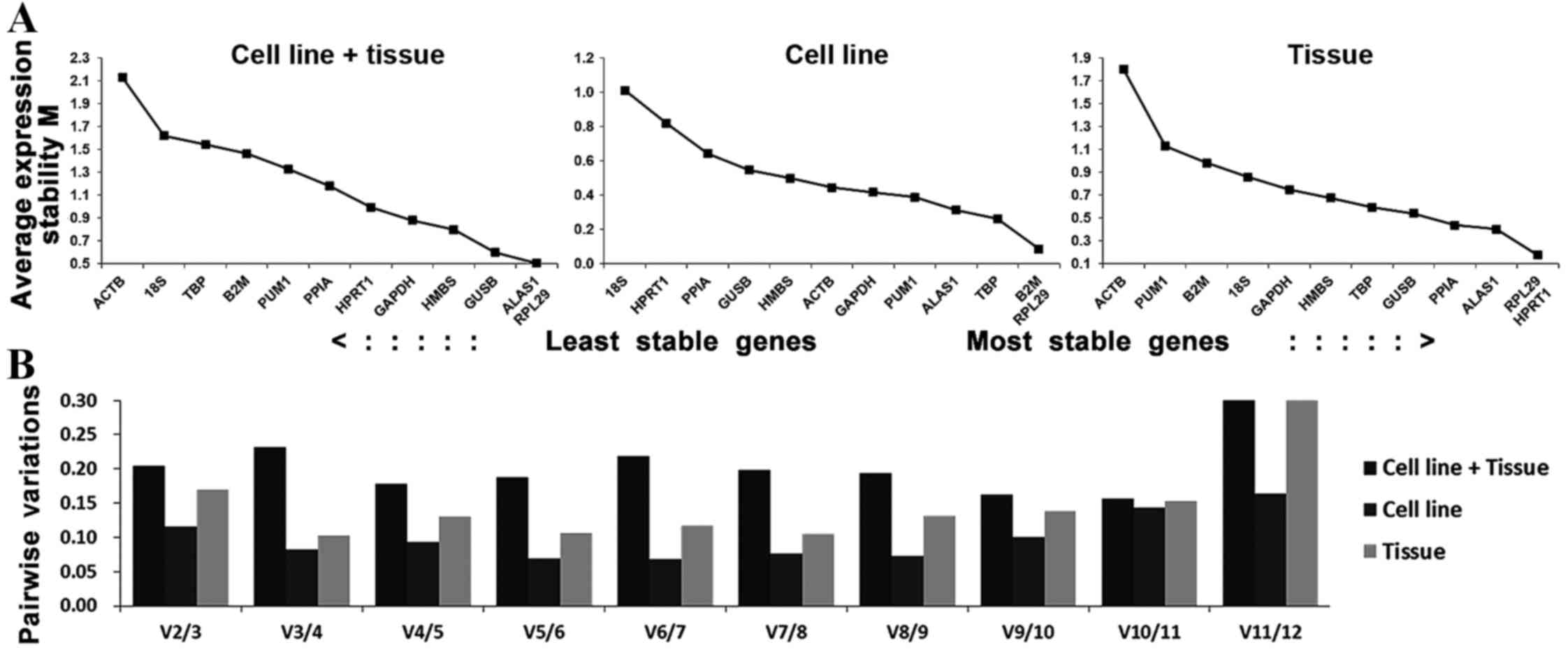
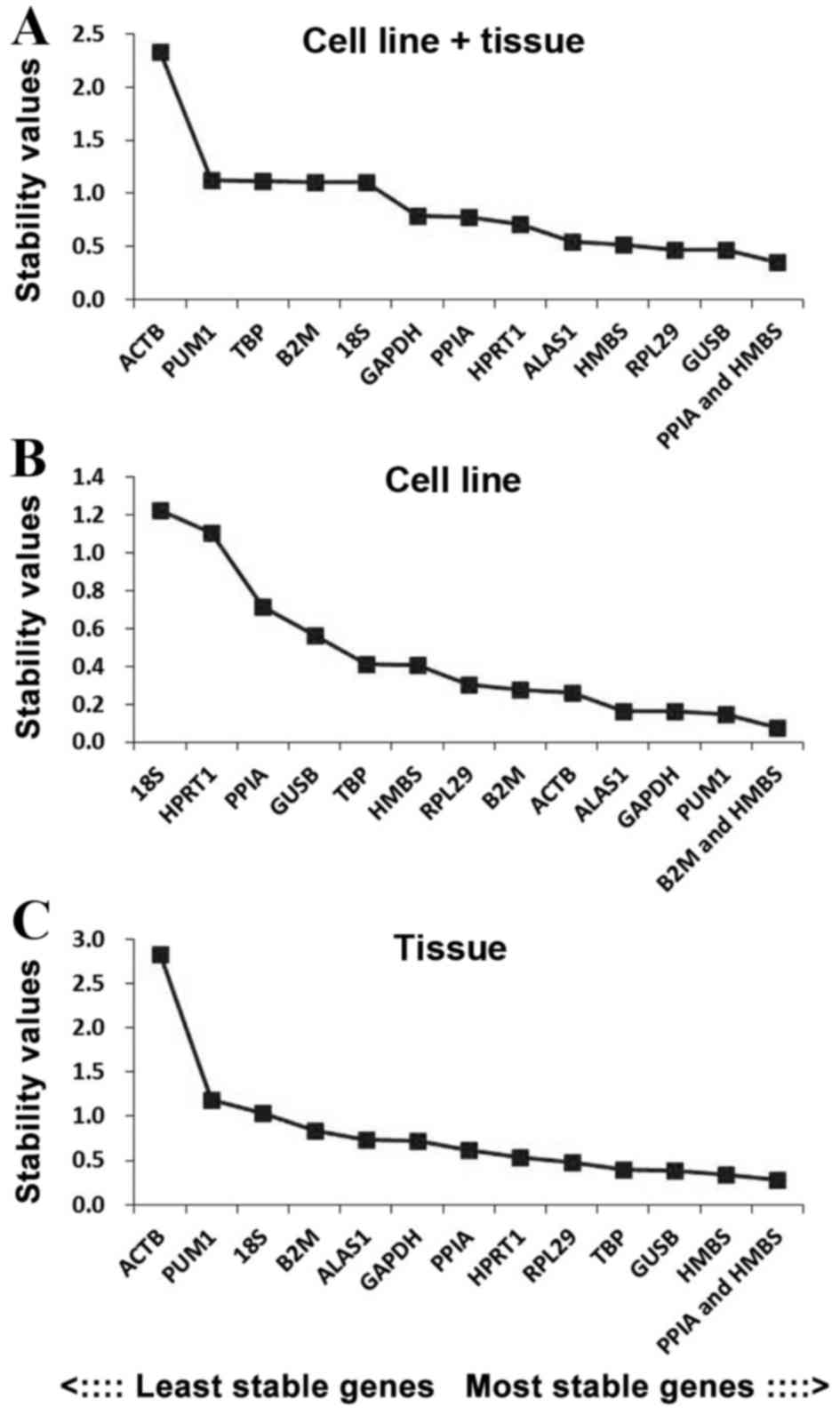
|
1
|
Radonić A, Thulke S, Mackay IM, Landt O,
Siegert W and Nitsche A: Guideline to reference gene selection for
quantitative real-time PCR. Biochem Biophys Res Commun.
313:856–862. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Derveaux S, Vandesompele J and Hellemans
J: How to do successful gene expression analysis using real-time
PCR. Methods. 50:227–230. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ali H, Du Z, Li X, Yang Q, Zhang YC, Wu M,
Li Y and Zhang G: Identification of suitable reference genes for
gene expression studies using quantitative polymerase chain
reaction in lung cancer in vitro. Mol Med Rep. 11:3767–3773.
2015.PubMed/NCBI
|
|
4
|
Ma H, Yang Q, Li D and Liu J: Validation
of suitable reference genes for quantitative polymerase chain
reaction analysis in rabbit bone marrow mesenchymal stem cell
differentiation. Mol Med Rep. 12:2961–2968. 2015.PubMed/NCBI
|
|
5
|
Yang Q, Ali HA, Yu S, Zhang L, Li X, Du Z
and Zhang G: Evaluation and validation of the suitable control
genes for quantitative PCR studies in plasma DNA for non-invasive
prenatal diagnosis. Int J Mol Med. 34:1681–1687. 2014.PubMed/NCBI
|
|
6
|
Yu S, Yang Q, Yang JH, Du Z and Zhang G:
Identification of suitable reference genes for investigating gene
expression in human gallbladder carcinoma using reverse
transcription quantitative polymerase chain reaction. Mol Med Rep.
11:2967–2974. 2015.PubMed/NCBI
|
|
7
|
He YX, Zhang Y, Yang Q, Wang C and Su G:
Selection of suitable reference genes for reverse
transcription-quantitative polymerase chain reaction analysis of
neuronal cells differentiated from bone mesenchymal stem cells. Mol
Med Rep. 12:2291–2300. 2015.PubMed/NCBI
|
|
8
|
Li X, Yang Q, Bai J, Xuan Y and Wang Y:
Identification of appropriate reference genes for human mesenchymal
stem cell analysis by quantitative real-time PCR. Biotechnol Lett.
37:67–73. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Li X, Yang Q, Bai J, Yang Y, Zhong L and
Wang Y: Identification of optimal reference genes for quantitative
PCR studies on human mesenchymal stem cells. Mol Med Rep.
11:1304–1311. 2015.PubMed/NCBI
|
|
10
|
Moore SR, Johnson NW, Pierce AM and Wilson
DF: The epidemiology of tongue cancer: A review of global
incidence. Oral Dis. 6:75–84. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhang S, Du Y, Tao J, Wu Y and Chen N:
Expression of cytosolic phospholipase A2 and cyclooxygenase 2 and
their significance in human oral mucosae, dysplasias and squamous
cell carcinomas. ORL J Otorhinolaryngol Relat Spec. 70:242–248.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ohl F, Jung M, Xu C, Stephan C, Rabien A,
Burkhardt M, Nitsche A, Kristiansen G, Loening SA, Radonić A and
Jung K: Gene expression studies in prostate cancer tissue: Which
reference gene should be selected for normalization? J Mol Med
(Berl). 83:1014–1024. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Huan P, Maosheng T, Zhiqian H, Long C and
Xiaojun Y: TLR4 expression in normal gallbladder, chronic
cholecystitis and gallbladder carcinoma. Hepatogastroenterology.
59:42–46. 2012.PubMed/NCBI
|
|
14
|
Vandesompele J, De Preter K, Pattyn F,
Poppe B, Van Roy N, De Paepe A and Speleman F: Accurate
normalization of real-time quantitative RT-PCR data by geometric
averaging of multiple internal control genes. Genome Biol.
3:Research00342002. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Andersen CL, Jensen JL and Ørntoft TF:
Normalization of real-time quantitative reverse transcription-PCR
data: A model-based variance estimation approach to identify genes
suited for normalization, applied to bladder and colon cancer data
sets. Cancer Res. 64:5245–5250. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Pfaffl MW, Tichopad A, Prgomet C and
Neuvians TP: Determination of stable housekeeping genes,
differentially regulated target genes and sample integrity:
BestKeeper-Excel-based tool using pair-wise correlations.
Biotechnol Lett. 26:509–515. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Sobin LH, Gospodarowicz MK and Wittekind
C: International Union Against CancerTNM Classification of
Malignant Tumours. 7th. Wiley-Blackwell; Chichester, UK: 2009
|
|
18
|
Battula VL, Bareiss PM, Treml S, Conrad S,
Albert I, Hojak S, Abele H, Schewe B, Just L, Skutella T and
Bühring HJ: Human placenta and bone marrow derived MSC cultured in
serum-free, b-FGF-containing medium express cell surface frizzled-9
and SSEA-4 and give rise to multilineage differentiation.
Differentiation. 75:279–291. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Mane VP, Heuer MA, Hillyer P, Navarro MB
and Rabin RL: Systematic method for determining an ideal
housekeeping gene for real-time PCR analysis. J Biomol Tech.
19:342–347. 2008.PubMed/NCBI
|
|
20
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wan H, Zhao Z, Qian C, Sui Y, Malik AA and
Chen J: Selection of appropriate reference genes for gene
expression studies by quantitative real-time polymerase chain
reaction in cucumber. Anal Biochem. 399:257–261. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Chang E, Shi S, Liu J, Cheng T, Xue L,
Yang X, Yang W, Lan Q and Jiang Z: Selection of reference genes for
quantitative gene expression studies in Platycladus orientalis
(Cupressaceae) Using real-time PCR. PLoS One. 7:e332782012.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Brugè F, Venditti E, Tiano L, Littarru GP
and Damiani E: Reference gene validation for qPCR on normoxia- and
hypoxia-cultured human dermal fibroblasts exposed to UVA: Is
β-actin a reliable normalizer for photoaging studies? J Biotechnol.
156:153–162. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wisnieski F, Calcagno DQ, Leal MF, dos
Santos LC, Gigek Cde O, Chen ES, Pontes TB, Assumpção PP, de
Assumpção MB, Demachki S, et al: Reference genes for quantitative
RT-PCR data in gastric tissues and cell lines. World J
Gastroenterol. 19:7121–7128. 2013. View Article : Google Scholar : PubMed/NCBI
|