Introduction
Primary human hepatocellular carcinoma (HCC) is the
most common form of liver cancer, the fifth most frequently
diagnosed type of malignancy and the third leading cause of
cancer-associated mortality, worldwide (1). According to statistical analysis, in
2015 an estimated 35,660 novel cases of HCC and 24,550
HCC-associated mortalities were recorded in the USA (2). Chronic hepatitis B virus (HBV) or
hepatitis C virus infection, alcohol abuse, non-alcoholic fatty
liver disease, autoimmune mediated hepatitis, primary biliary
cirrhosis and exposure to carcinogens are the primary risk factors
of HCC (3). Among these, infection
with HBV is the most important risk factor associated with the
disease worldwide and is responsible for the increasing incidence
of HCC, particularly in China (4,5). Patients
with HCC from China comprise 55% of all patients with HCC worldwide
(6). Currently, liver resection,
liver transplantation, radiotherapy, chemotherapy and targeted
therapy are the standard therapeutic treatments used for patients
with early stage HCC (7,8). However, the 5-year survival rate remains
low, particularly for patients with HCC who are diagnosed at
advanced stages, mainly due to the lack of effective therapeutic
treatments (4,9). Therefore, it is important to investigate
novel therapeutic approaches in order to improve the effectiveness
of treatment and the prognosis for patients with HCC.
MicroRNAs (miRNAs/miR) belong to a group of
evolutionarily conserved and non-coding small RNAs, 19–25
nucleotides in length (10). In
humans, 1,881 miRNA precursors and 2,588 mature miRNA sequences
have been deposited in miRBase since the first discovery of miRNA
lin-4 in Caenorhabditis elegans (11–13). They
have been demonstrated to negatively modulate the expression of
targeted mRNAs in animals, plants and viruses by directly binding
to the 3′untranslated region (3′UTR) of targeted mRNAs in base
pairing, resulting in mRNA degradation or translational inhibition
at the translational or post-transcriptional levels (14,15).
Numerous previous studies have demonstrated that miRNAs are
involved in a variety of cellular biological processes, including
proliferation, the cell cycle, apoptosis, invasion, migration and
metastasis (16–18). The abnormal expression levels of
miRNAs have been identified in various types of human malignancies,
including HCC, suggesting that miRNAs may contribute to the
carcinogenesis and progression of these types of cancer (19–21).
Previous studies have also indicated that miRNAs may be potential
novel therapeutic targets for a number of cancer subtypes (22,23).
Therefore, it is important to further study the abnormally
expressed miRNAs and their roles in HCC, which may provide
effective and novel therapeutic targets for HCC. The expression
level and functions of miR-204 have been studied in several types
of cancer; however, the role of miR-204 in HCC has yet to be
elucidated. The present study aimed to investigate the expression
patterns, biological functions and underlying molecular mechanisms
of miR-204 in HCC.
Materials and methods
Clinical specimens and cell lines
A total of 82 HCC tissues and paired adjacent
non-tumorous liver tissues were used in the present retrospective
study. These were obtained from 82 HCC patients (including 34 male
and 58 female patients; age range, 38–72 years) who were treated
with surgical resection at The First Affiliated Hospital of Xiamen
University (Xiamen, China) between May 2013 and March 2015. All of
the patients enrolled in the study did not receive chemotherapy,
radiotherapy or other treatment prior to surgery. Patients treated
with chemotherapy or radiotherapy were excluded form the present
study. HCC tissues and non-tumorous liver tissues were snap-frozen
in liquid nitrogen following surgery and stored at −80°C. The
present study was approved by the Research Ethics Committee of The
First Affiliated Hospital of Xiamen University (Xiamen, China).
Written informed consent was obtained from all patients prior to
enrollment in the present study.
The human HCC cell lines (HepG2, PLC-5, SMMC-7721,
HuH-7 and HLE), and the immortalized normal liver epithelial cell
line (THLE-3) were purchased from the American Type Culture
Collection (Manassas, VA, USA) The cells were cultured in high
glucose Dulbecco's modified Eagle's medium (Gibco; Thermo Fisher
Scientific, Inc., Waltham, MA, USA) supplemented with 10% v/v fetal
bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.), 100
IU/ml penicillin and 100 µg/ml streptomycin (Gibco; Thermo Fisher
Scientific, Inc.).
Cell transfection
miR-204 mimics and the corresponding negative
control (NC) were purchased from GenePharma, Inc. (Sunnyvale, CA,
USA). The pGL3-ZEB2-3′UTR wild-type (Wt) and pGL3-ZEB2-3′UTR mutant
(Mut) were also obtained from GenePharma, Inc. For the knockdown of
ZEB2 expression, small interfering (si) RNAs targeting ZEB2 or NC
siRNA were purchased from Guangzhou Ribobio Co., Ltd. (Guangzhou,
China). Oligonucleotide transfection or co-transfection was
performed using Lipofectamine® 2000 (Invitrogen; Thermo
Fisher Scientific, Inc.), according to the manufacturer's
instructions.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was isolated from HCC tissues, adjacent
non-tumor liver tissues, HCC cell lines and THLE-3 immortalized
normal liver epithelial cells using TRIzol® (Invitrogen;
Thermo Fisher Scientific, Inc.), following the manufacturer's
protocol. In order to evaluate the expression profile of miR-204,
complementary DNA (cDNA) was synthesized from 1 µg of total RNA,
using the TaqMan® MicroRNA Reverse Transcription kit
(Applied Biosystems; Thermo Fisher Scientific, Inc.), followed by
qPCR using the specific TaqMan MicroRNA assay (Applied Biosystems;
Thermo Fisher Scientific, Inc.).
In order to determine the mRNA expression level of
ZEB2, cDNA was synthesized using the GeneAmp RNA PCR kit (Thermo
Fisher Scientific, Inc.), followed by qPCR using the Power
SYBR® Green Master Mix (Thermo Fisher Scientific, Inc.).
The RT-qPCR process was performed using the Applied Biosystems 7500
Real-time PCR System (Applied Biosystems; Thermo Fisher Scientific,
Inc.). For both qPCR procedures, the thermocycling conditions were
as follows: 95°C for 10 min, then 40 cycles of 95°C for 15 sec and
60°C for 1 min.
The primer sequences for miR-204 were as follows:
Forward, 5′-GCGGCGCAAAGAATTCTCCT-3′, reverse,
5′-GTGCAGGGTCCGAGGT-3′; U6 forward, 5′-CTCGCTTCGGCAGCACA-3′,
reverse, 5′-AACGCTTCACGAATTTGCGT-3′; ZEB2 forward,
5′-GCAATGTAGGTCTCTGCTGC-3′, reverse, 5′-CTCCCCTTTGCTCCTTCTCA-3′;
GAPDH forward, 5′-CGGAGTCAACGGATTTGGTCGTAT-3′, reverse,
5′-AGCCTTCTCCATGGTGGTGAAGAC-3′. U6 and GADPH were used as internal
controls for miR-204 and ZEB2 mRNA expression, respectively. The
relative expression level of miR-294 and ZEB2 mRNA was determined
with the 2−ΔΔCq method (24).
Bioinformatics analysis
In order to find putative targets of miR-204, a
bioinformatics analysis was performed with TargetScan (http://www.targetscan.org/).
Dual-luciferase reporter assay
For the reporter assays, cells were seeded into
24-well plates at 50–60% confluence, and co-transfected with
miR-204 mimics (20 pmol), NC (20 pmol) and pGL3-ZEB2-3′UTR Wt (1
µg) or pGL3-ZEB2-3′UTR Mut (1 µg) at room temperature. Following a
48-h transfection, cells were collected and the luciferase
activities were determined by performing a dual-luciferase reporter
assay (Promega Corporation, Madison, WI, USA), according to the
manufacturer's protocol. The Renilla luciferase activities
were measured using an xMark™ microplate absorbance
spectrophotometer (Bio-Rad Laboratories, Inc.) and used as internal
controls for firefly luciferase activity.
Western blot analysis
Transfected HepG2 and HuH-7 cells were washed with
PBS, harvested with a cell scraper and homogenized using
radioimmunoprecipitation assay lysis buffer (Beyotime Institute of
Biotechnology, Haimen, China) supplemented with protease inhibitor
(Pierce; Thermo Fisher Scientific, Inc.), according to the
manufacturer's protocol. The protein concentration was detected
using a bicinchoninic acid assay protein kit (Thermo Fisher
Scientific, Inc.), and equal amounts of protein (20 µg) were
separated using 10% SDS-PAGE and then transferred to a
polyvinylidine fluoride membrane (Invitrogen; Thermo Fisher
Scientific, Inc.). Subsequently, the membranes were blocked with
TBS and Tween-20 (TBST) supplemented with 5% skimmed milk for 2 h
at room temperature prior to incubation with primary antibodies
overnight at 4°C. The primary antibodies used were mouse anti-human
monoclonal ZEB2 (1:1,000 dilution; cat. no. sc-271984; Santa Cruz
Biotechnology, Inc., Dallas, TX, USA) and mouse anti-human
monoclonal β-actin (1:1,000 dilution; cat. no. sc-47778; Santa Cruz
Biotechnology, Inc.). β-actin was used as the internal control for
ZEB2. Following washing with TBST three times, the membranes were
probed with goat anti-mouse IgG horseradish peroxidase-conjugated
secondary antibody (1:5,000 dilution; cat. no. sc-2005; Santa Cruz
Biotechnology, Inc.) at room temperature for 2 h and visualized
using a FluorChem imaging system (Alpha Innotech Co., San Leandro,
CA, USA).
Cell proliferation assay
Cell counting kit-8 (CCK8; Dojindo Molecular
Technologies, Inc., Kumamoto, Japan) was used to determine the
effect of miR-204 on the proliferative ability of cells. Cells were
seeded into 96-well plates at a density of 3,000 cells/well.
Following overnight incubation at 37°C with 5% CO2,
cells were transfected with miRNAs (50 pmol/ml) or siRNAs (50
pmol/ml). A CCK8 assay was performed every 24 h subsequent to
transfection, up to a total of 96 h. In brief, cells were treated
with 10 µl CCK8 solution for 2 h at 37°C. Absorbance was detected
at 450 nm for each well using a microplate reader (Bio-Rad
Laboratories, Inc., Hercules, CA, USA). All experiments were
performed with 5 replications.
Cell migration and invasion
assays
Cell migration and invasion assays were performed
with a Transwell chambers (8-µm pore size; Corning Incorporated,
Corning, NY, USA) coated with (invasion assay) or without
(migration assay) Matrigel (BD Biosciences, San Jose, CA, USA). For
migration and invasion assays, 1×105 transfected cells
in 300 µl FBS-free culture medium were seeded into the upper
Transwell chambers. The lower Transwell chambers received 500 µl
culture medium supplemented with 20% FBS. Following a 24-h
incubation at 37°C with 5% CO2, non-migrated cells were
removed using a cotton swab. Cells migrating or invading to the
lower surface membranes of the Transwell chambers were fixed with
methanol and stained with 0.5% crystal violet. Subsequent to
washing with PBS three times, cells were counted in five randomly
selected visual fields under an inverted microscope (magnification,
×200; Olympus Corporation, Tokyo, Japan).
Statistical analysis
Data are expressed as the mean ± standard deviation,
and were analyzed using Student's t-test or one-way analysis
of variance, followed by Student Newman Keuls post hoc test, with
SPSS 19.0 software (IBM SPSS, Armonk, NY, USA). P<0.05 was
considered to indicate a statistically significant difference.
Results
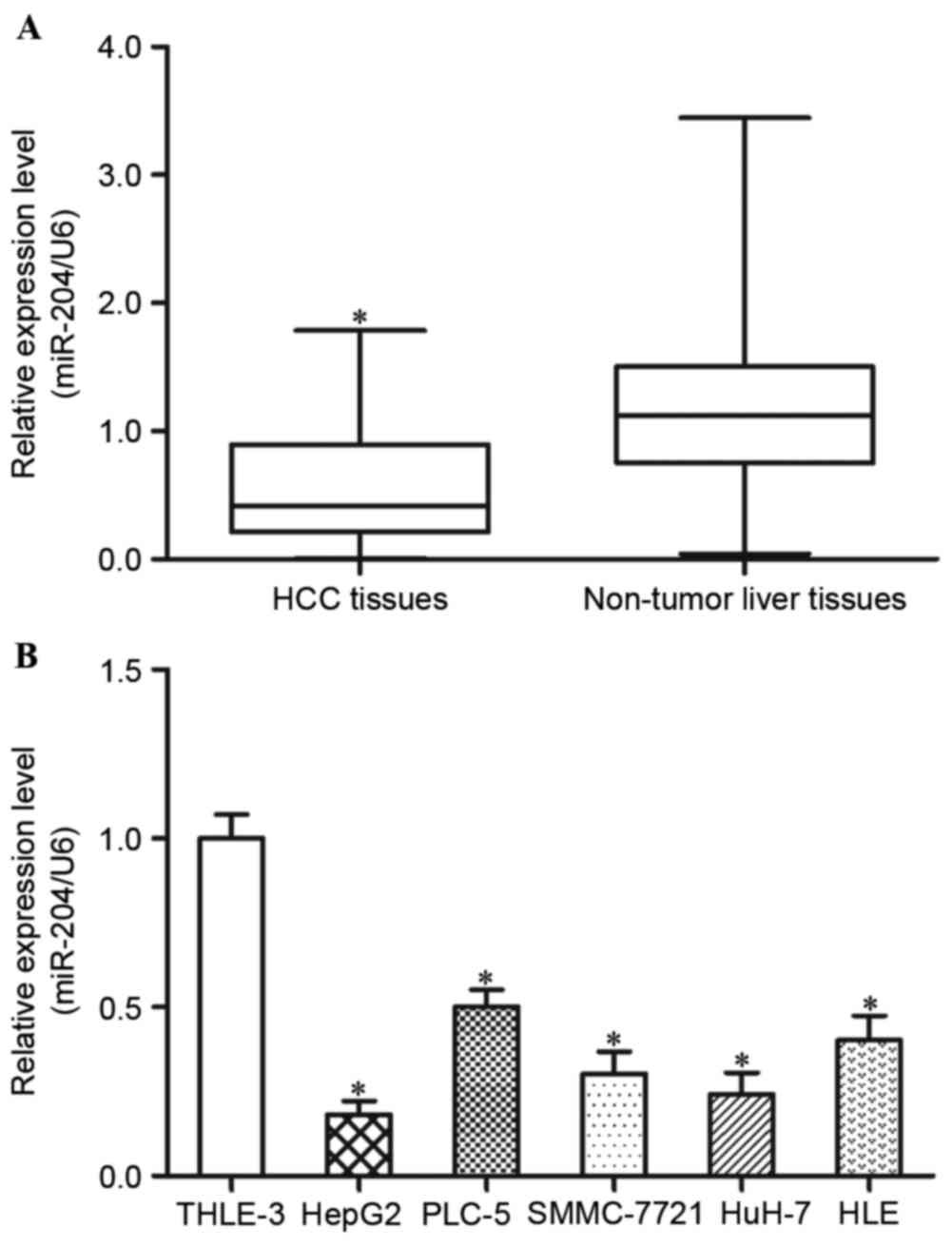
miR-204 is downregulated in HCC
tissues and cell lines
In order to evaluate the expression levels of
miR-204 in HCC tissues, RT-qPCR was performed to determine the
differential expression levels of miR-204 in HCC tissues and paired
adjacent non-tumorous liver tissues. The results demonstrated that
miR-204 expression levels are decreased in HCC tissues compared
with in adjacent non-tumorous liver tissues (Fig. 1A, P<0.05).
In addition, the present study detected the
expression levels of miR-204 in five HCC cell lines (HepG2, PLC-5,
SMMC-7721, HuH-7 and HLE) and an immortalized normal liver
epithelial cell line (THLE-3). The results demonstrated that,
compared with THLE-3 cells, the five HCC cell lines exhibited lower
expression levels of miR-204 (Fig.
1B, P<0.05). Within the five HCC cell lines, HepG2 and HuH-7
revealed the lowest miR-204 expression levels, and were selected
for further functional studies. These findings indicated that
miR-204 may be involved in the carcinogenesis and progression of
HCC.
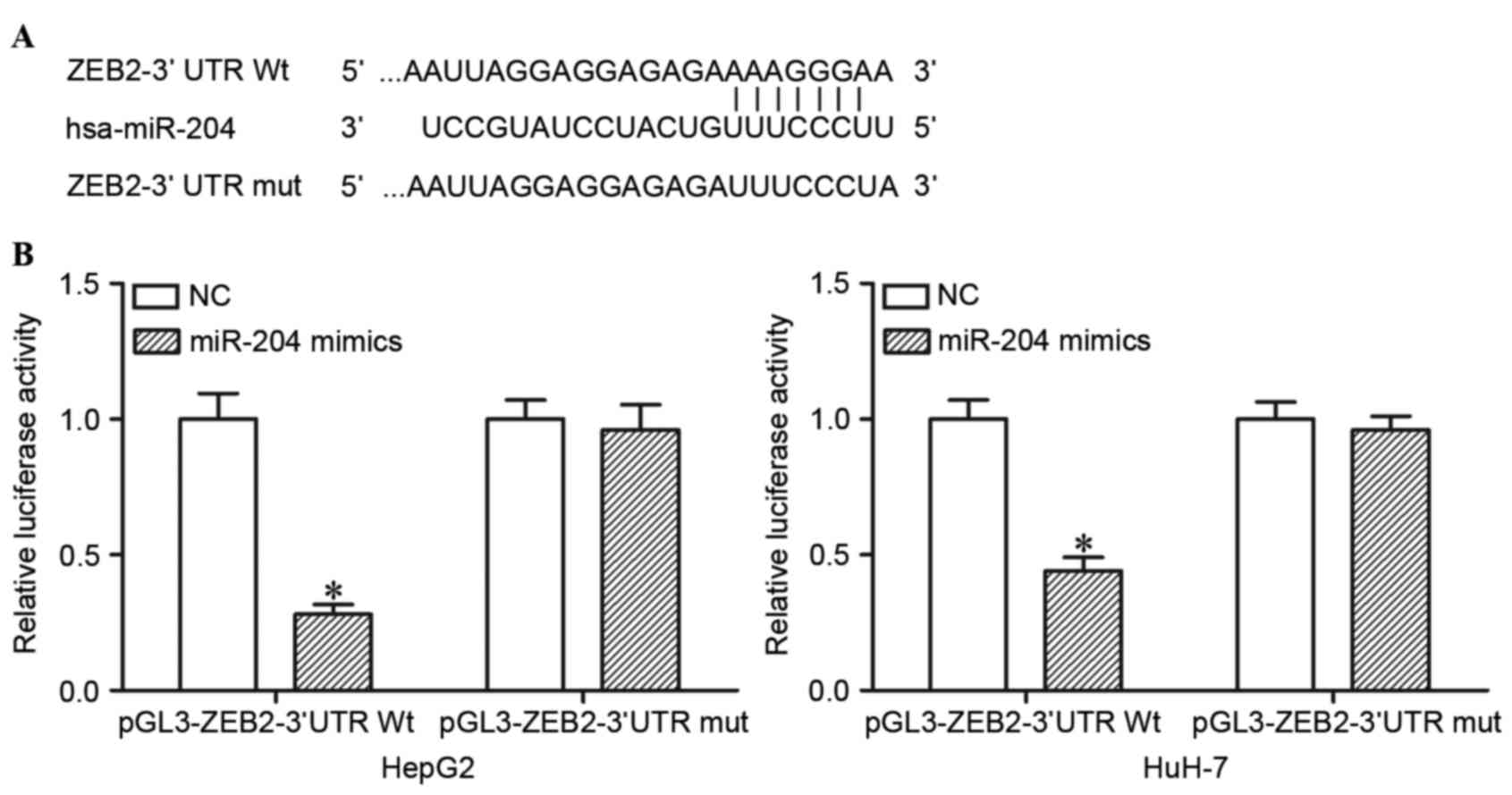
miR-204 directly targets the 3′UTR of
ZEB2 in HCC cells
TargetScan was used to predict the potential target
genes of miR-204. The analysis revealed that ZEB2 contained a
miR-204 seed match at position 218–225 of ZEB2 3′UTR (Fig. 2A). In order to confirm whether miR-204
is a direct target of miR-204, dual luciferase reporter assays were
performed. As presented in Fig. 2B,
ectopic miR-204 expression in HepG2 and HuH-7 cells significantly
suppressed the luciferase activities of pGL3-ZEB2-3′UTR Wt
(P<0.05), but had no observable effect on the luciferase
activities of pGL3- ZEB2-3′UTR Mut. miR-204 directly targeted the
3′UTR of ZEB2 in HCC cells.
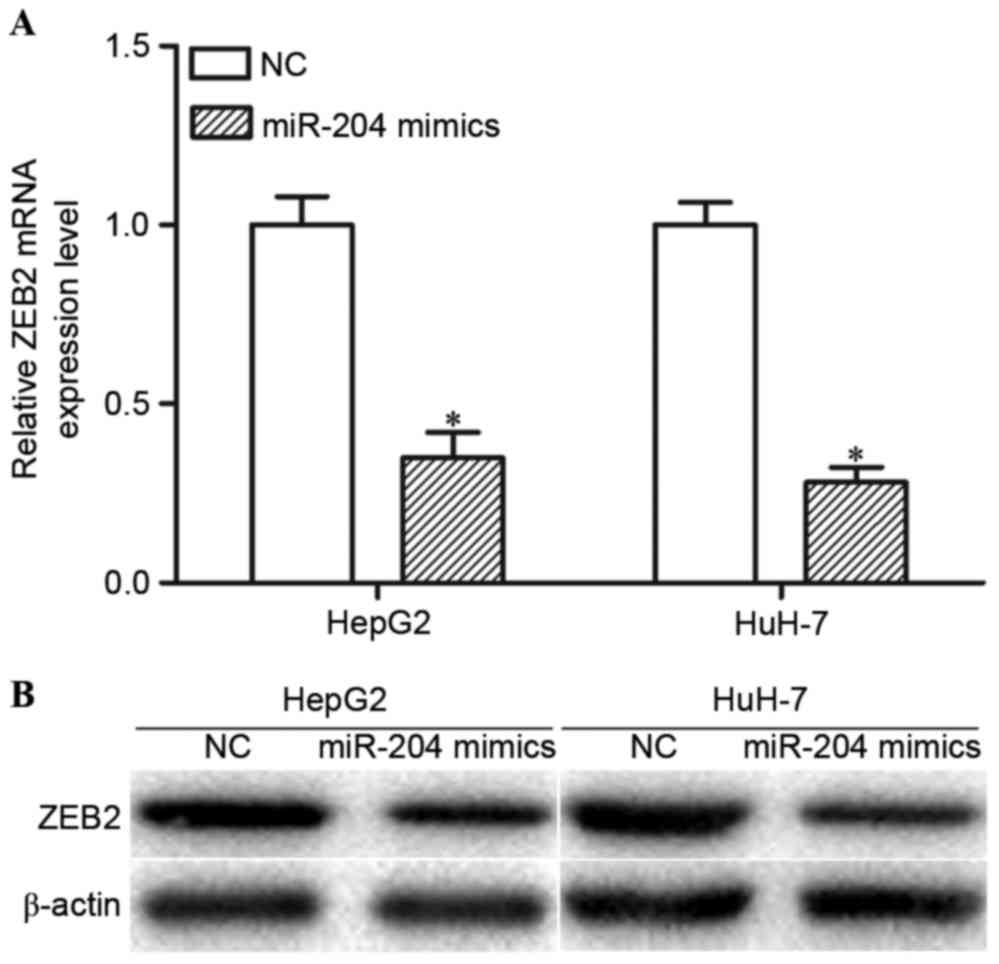
miR-204 negatively regulates ZEB2
expression levels at the post-transcriptional level
The present study performed RT-qPCR and western blot
analysis in order to determine whether miR-204 regulated ZEB2
expression. The RT-qPCR results indicated that high expression
levels of miR-204 decreased ZEB2 mRNA expression levels in HepG2
and HuH-7 cells (Fig. 3A; P<0.05).
Western blot analysis revealed that overexpression of miR-204 may
reduce ZEB2 protein expression levels in HepG2 and HuH-7 cells
(Fig. 3B; P<0.05). Collectively,
miR-204 negatively modulated ZEB2 expression levels in HCC cells
via an underlying mRNA cleavage mechanism at the
post-transcriptional level.
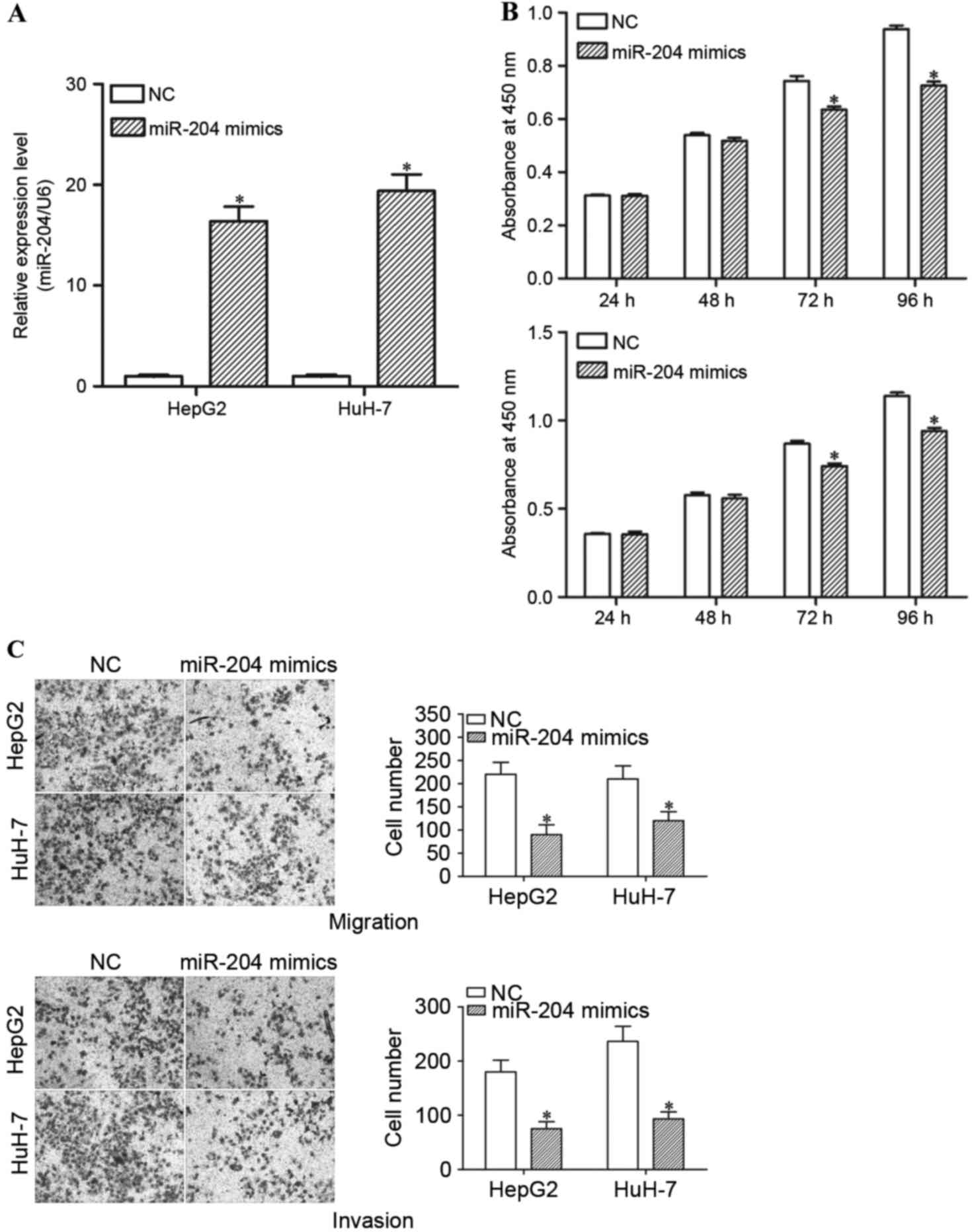
miR-204 suppresses the proliferation,
migration and invasion of HCC cells
In order to investigate the function of miR-204 in
HCC cells, miR-204 mimics or NCs were introduced into HepG2 and
HuH-7 cells. RT-qPCR was performed to determine the transfection
efficiency. The results demonstrated that miR-204 expression levels
are markedly increased in miR-204 mimic-transfected HepG2 and HuH-7
cells, compared with the NC groups (Fig.
4A; P<0.05).
Subsequently, cell proliferation, cell migration and
invasion assays were performed in order to investigate the
functions of miR-204 in the growth, migration and invasion of HCC
cells. The results demonstrated that high expression levels of
miR-204 resulted in a significant decrease in the cell
proliferation of HepG2 and HuH-7 cells (Fig. 4B; P<0.05). The migration and
invasion assays revealed that enforced miR-204 expression
suppresses the migration and invasion ability of HepG2 and HuH-7
cells (Fig. 4C; P<0.05). Thus,
restoration of miR-204 expression suppresses growth and motility in
HCC.
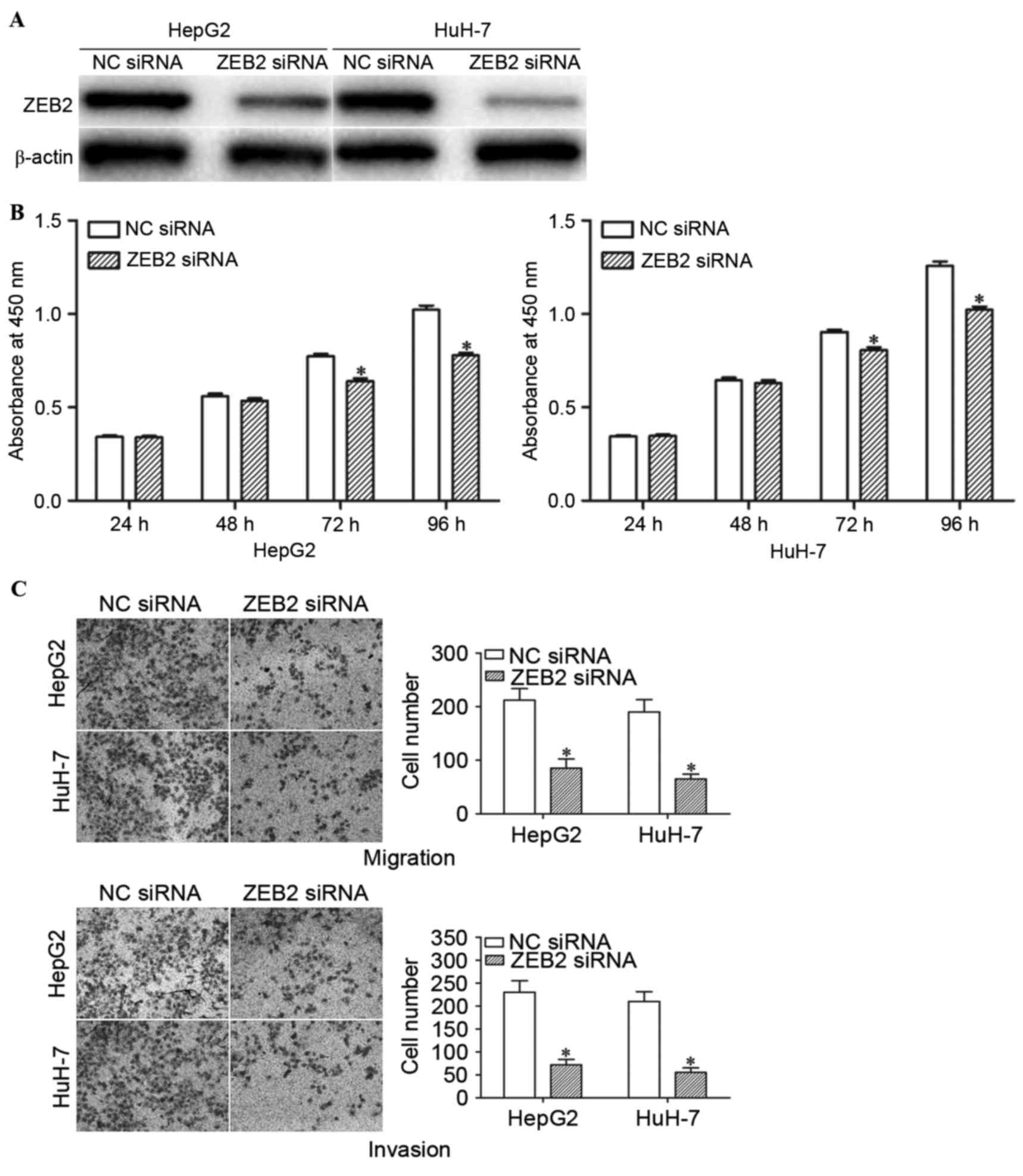
Knockdown of ZEB2 inhibits the
proliferation, migration and invasion of HCC cells
In order to determine whether the functions of
miR-204 in HCC cells are mediated by ZEB2, HepG2 and HuH-7 cells
were transfected with ZEB2 siRNA or NC siRNA. Western blot analysis
confirmed that ZEB2 siRNA significantly decreased the expression
levels of ZEB2 in HepG2 and HuH-7 cells (Fig. 5A; P<0.05).
The effects of ZEB2 siRNA on the proliferation,
migration and invasion of HCC cells were determined using cell
proliferation, cell migration and invasion assays. Consistently,
ZEB2 siRNA suppressed the proliferation, migration and invasion of
HepG2 and HuH-7 cells (Fig. 5B and C;
P<0.05). These results indicate that miR-204 inhibit the growth,
migration and invasion of HCC cells via knockdown of ZEB2
expression.
Discussion
HCC is one of the most common malignant types of
cancer worldwide and the second most common cause of
cancer-associated mortality in China (25). Therefore, understanding the molecular
mechanisms underlying the carcinogenesis and progression of HCC is
required for the development of novel therapeutic treatments for
patients with HCC, in order to improve the rate of survival. A
number of previous studies have demonstrated that abnormal
expression levels of miRNAs may be important in HCC initiation and
development, suggesting that miRNA may be investigated as a novel
direction in the targeted therapy of HCC (26–28).
The present study initially demonstrated that
miR-204 was significantly downregulated in HCC tissues and cell
lines, indicating that abnormal expression levels of miR-204 may
contribute to HCC initiation and development. In addition,
TargetScan was used to predict the potential target genes of
miR-204. The results revealed that ZEB2 contains a miR-204 seed
match at its 3′UTR position. Dual luciferase reporter assays
indicated that miR-204 directly targets the 3′UTR of ZEB2. RT-qPCR
and western blot analysis demonstrated that ZEB2 mRNA and protein
expression levels are decreased in HCC cells transfected with
miR-204 mimics. These results indicated that miR-204 negatively
regulates ZEB2 expression levels at the post transcription level
via directly targeting the 3′UTR of ZEB2. Furthermore, the present
study determined the rates of cell growth and metastasis by
performing cell proliferation, cell migration and invasion assays,
in order to evaluate the association between miR-204 expression and
the growth and metastasis capacity of HCC cells. The cell growth,
viability and metastasis of HCC cells transfected with the miR-204
mimics was decreased, compared with in the NC groups. Finally, the
functions of ZEB2 in HCC cells were determined. The results
revealed that the downregulation of ZEB2 may mimic the functions of
miR-204 in HCC cells, suggesting that ZEB2 is a functional target
of miR-204 in HCC cells.
miR-204 was previously revealed to be downregulated
in a number of human tumor subtypes, including thyroid cancer
(29), renal cell carcinoma (30), breast cancer (31), glioma (32), acute myeloid leukemia (33), osteosarcoma (34) and ovarian cancer (35). In functional studies, miR-204 was
demonstrated to be a tumor suppressor in the aforementioned types
of cancer. For example, in thyroid cancer, miR-204 suppressed cell
proliferation by targeting high-mobility group AT-hook 2 and
insulin like growth factor binding protein 5 (29,36). Wu
et al (30) reported that
overexpressed miR-204 targeted SRY-Box 4 in order to inhibit the
growth, migration and invasion of renal cell carcinoma cells. In
breast cancer, low expression levels of miR-204 were revealed to be
associated with the tumor-node-metastasis stage, metastasis and
chemotherapeutic resistance of patients with breast cancer. In
addition, patients with low miR-204 expression levels demonstrated
a poorer overall survival time and disease free survival time,
compared with those with high miR-204 expression levels (37). miR-204 increased the extent of
apoptosis in breast cancer cells via targeting Janus kinase 2
through the signal transducer and activator of transcription
3/B-cell lymphoma 2 signaling pathway (31). Xia et al (32) demonstrated that the upregulation of
miR-204 significantly decreased glioma cell growth, migration and
invasion in vitro, and suppressed tumorigenesis and
increased the overall patient survival rate in vivo by the
regulation of RAB22A. Mao et al (38) reported that miR-204 targeted ezrin in
order to decrease the migration and invasion abilities of glioma.
These findings collectively suggest that miR-204 serves important
roles in tumor suppression, and should be investigated as a
potential targeted therapeutic treatment for the aforementioned
types of cancer.
Identification of miR-204 target mRNAs is required
for investigating its functions in carcinogenesis and the
progression of HCC, and for the development of novel targeted
therapies (39). In the present
study, ZEB2 was identified as a functional target gene of miR-204
in HCC cells. ZEB2 is a member of the zinc finger family, and has
previously demonstrated upregulation in a number of human cancer
subtypes, including breast cancer, gastric cancer, glioma, ovarian
cancer and non-small cell lung carcinoma (40–46). ZEB2
has also been demonstrated to be upregulated in HCC, and expression
levels of ZEB2 in the nucleus were associated with angiogenesis and
metastasis. Patients with HCC and ZEB2 nuclear expression exhibited
shorter survival times compared with those without ZEB2 expression.
In previous studies, the overexpression of ZEB2 induced motility,
invasiveness and angiogenesis of HCC cells (47). By contrast, the knockdown of ZEB2
suppressed HCC cell motility, invasiveness and vasculogenic mimicry
formation (47). This was in
accordance with the results of the present study.
It has previously been demonstrated that ZEB2 was
regulated by numerous miRNAs in a number of human cancer subtypes.
For example, in gastric cancer, miR-141 targeted ZEB2 in order to
inhibit cancer cell migration (48).
Zhou et al (49) reported that
miR-153 suppressed cell growth and invasion of ovarian cancer by
directly targeting ZEB2. In renal cell carcinoma, miR-205 inhibited
cancer cell growth, migration, invasion and induced apoptosis via
blockade of ZEB2 (50). These studies
indicated that miRNAs may serve as a regulator of ZEB2 in human
types of cancer. In the present study, the upregulation of miR-204
in HCC cell lines demonstrated that miR-204 inhibits cell growth,
migration and invasion by negatively regulating ZEB2. Therefore,
miR-204 maybe investigated as a possible targeted therapy for
HCC.
In conclusion, the expression of miR-204 was
downregulated in HCC tissues and cell lines. In vitro
functional studies revealed that miR-204 acted as a tumor
suppressor by inhibiting the proliferation, migration and invasion
of HCC. Its numerous tumor suppressor functions are mediated by
ZEB2. The downregulation of miR-204 may serve an important role in
tumor growth and metastasis, and may be a potential therapeutic
target for HCC.
Acknowledgements
The present study was supported by the National
Natural Science Foundation of China (grant nos. 81371902 and
81301923) and the National Natural Science Foundation of Fujian
(grant nos. 2011D012, 2015J01561 and 2016J01633).
References
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2015. CA Cancer J Clin. 65:5–29. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Sanyal AJ, Yoon SK and Lencioni R: The
etiology of hepatocellular carcinoma and consequences for
treatment. Oncologist. 15:(Suppl 4). 14–22. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
El-Serag HB and Rudolph KL: Hepatocellular
carcinoma: Epidemiology and molecular carcinogenesis.
Gastroenterology. 132:2557–2576. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
El-Serag HB: Hepatocellular carcinoma. N
Engl J Med. 365:1118–1127. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Asia-Pacific Working Party on Prevention
of Hepatocellular Carcinoma: Prevention of hepatocellular carcinoma
in the Asia-Pacific region: consensus statements. J Gastroenterol
Hepatol. 25:657–663. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Liu D, Dong L, Liu Y, Wen D, Gao D, Sun H,
Fan J and Wu W: A c-Myc/miR-17-5p feedback loop regulates
metastasis and invasion of hepatocellular carcinoma. Tumour Biol.
37:5039–5047. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hsieh MY, Lin ZY, Chen SH and Chuang WL:
Risk factors for the leakage of chemotherapeutic agents into
systemic circulation after transcatheter arterial chemoembolization
of hepatocellular carcinoma. Kaohsiung J Med Sci. 27:431–436. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yang LY, Fang F, Ou DP, Wu W, Zeng ZJ and
Wu F: Solitary large hepatocellular carcinoma: A specific subtype
of hepatocellular carcinoma with good outcome after hepatic
resection. Ann Surg. 249:118–123. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Li Y and Kowdley KV: MicroRNAs in common
human diseases. Genomics Proteomics Bioinformatics. 10:246–253.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lee RC, Feinbaum RL and Ambros V: The C.
elegans heterochronic gene lin-4 encodes small RNAs with antisense
complementarity to lin-14. Cell. 75:843–854. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wightman B, Ha I and Ruvkun G:
Posttranscriptional regulation of the heterochronic gene lin-14 by
lin-4 mediates temporal pattern formation in C. elegans. Cell.
75:855–862. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kozomara A and Griffiths-Jones S: miRBase:
Integrating microRNA annotation and deep-sequencing data. Nucleic
Acids Res. 39:D152–D157. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
O'Hara SP, Mott JL, Splinter PL, Gores GJ
and LaRusso NF: MicroRNAs: Key modulators of posttranscriptional
gene expression. Gastroenterology. 136:17–25. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lujambio A and Lowe SW: The microcosmos of
cancer. Nature. 482:347–355. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Iorio MV and Croce CM: microRNA
involvement in human cancer. Carcinogenesis. 33:1126–1133. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ebert MS and Sharp PA: Roles for microRNAs
in conferring robustness to biological processes. Cell.
149:515–524. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yang N, Ekanem NR, Sakyi CA and Ray SD:
Hepatocellular carcinoma and microRNA: New perspectives on
therapeutics and diagnostics. Adv Drug Deliv Rev. 81:62–74. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Romero-Cordoba SL, Salido-Guadarrama I,
Rodriguez-Dorantes M and Hidalgo-Miranda A: miRNA biogenesis:
Biological impact in the development of cancer. Cancer Biol Ther.
15:1444–1455. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Lan H, Lu H, Wang X and Jin H: MicroRNAs
as potential biomarkers in cancer: Opportunities and challenges.
Biomed Res Int. 2015:1250942015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Petri A, Lindow M and Kauppinen S:
MicroRNA silencing in primates: Towards development of novel
therapeutics. Cancer Res. 69:393–395. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Landi MT, Zhao Y, Rotunno M, Koshiol J,
Liu H, Bergen AW, Rubagotti M, Goldstein AM, Linnoila I, Marincola
FM, et al: MicroRNA expression differentiates histology and
predicts survival of lung cancer. Clin Cancer Res. 16:430–441.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
He J, Gu D, Wu X, Reynolds K, Duan X, Yao
C, Wang J, Chen CS, Chen J, Wildman RP, et al: Major causes of
death among men and women in China. N Engl J Med. 353:1124–1134.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Chen X, Bo L, Lu W, Zhou G and Chen Q:
MicroRNA-148b targets Rho-associated protein kinase 1 to inhibit
cell proliferation, migration and invasion in hepatocellular
carcinoma. Mol Med Rep. 13:477–482. 2016.PubMed/NCBI
|
|
27
|
Chen X, Bo L, Zhao X and Chen Q:
MicroRNA-133a inhibits cell proliferation, colony formation
ability, migration and invasion by targeting matrix
metallopeptidase 9 in hepatocellular carcinoma. Mol Med Rep.
11:3900–3907. 2015.PubMed/NCBI
|
|
28
|
Pang X, Huang K, Zhang Q, Zhang Y and Niu
J: miR-154 targeting ZEB2 in hepatocellular carcinoma functions as
a potential tumor suppressor. Oncol Rep. 34:3272–3279.
2015.PubMed/NCBI
|
|
29
|
Wu ZY, Wang SM, Chen ZH, Huv SX, Huang K,
Huang BJ, Du JL, Huang CM, Peng L, Jian ZX and Zhao G: MiR-204
regulates HMGA2 expression and inhibits cell proliferation in human
thyroid cancer. Cancer Biomark. 15:535–542. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wu D, Pan H, Zhou Y, Zhang Z, Qu P, Zhou J
and Wang W: Upregulation of microRNA-204 inhibits cell
proliferation, migration and invasion in human renal cell carcinoma
cells by downregulating SOX4. Mol Med Rep. 12:7059–7064.
2015.PubMed/NCBI
|
|
31
|
Wang X, Qiu W, Zhang G, Xu S, Gao Q and
Yang Z: MicroRNA-204 targets JAK2 in breast cancer and induces cell
apoptosis through the STAT3/BCl-2/survivin pathway. Int J Clin Exp
Pathol. 8:5017–5025. 2015.PubMed/NCBI
|
|
32
|
Xia Z, Liu F, Zhang J and Liu L: Decreased
expression of MiRNA-204-5p contributes to glioma progression and
promotes glioma cell growth, migration and invasion. PLoS One.
10:e01323992015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Butrym A, Rybka J, Baczynska D, Tukiendorf
A, Kuliczkowski K and Mazur G: Low expression of microRNA-204
(miR-204) is associated with poor clinical outcome of acute myeloid
leukemia (AML) patients. J Exp Clin Cancer Res. 34:682015.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Shi Y, Huang J, Zhou J, Liu Y, Fu X, Li Y,
Yin G and Wen J: MicroRNA-204 inhibits proliferation, migration,
invasion and epithelial-mesenchymal transition in osteosarcoma
cells via targeting Sirtuin 1. Oncol Rep. 34:399–406.
2015.PubMed/NCBI
|
|
35
|
Yan H, Wu W, Ge H, Li P and Wang Z:
Up-regulation of miR-204 enhances anoikis sensitivity in epithelial
ovarian cancer cell line via brain-derived neurotrophic factor
pathway in vitro. Int J Gynecol Cancer. 25:944–952. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Liu L, Wang J, Li X, Ma J, Shi C, Zhu H,
Xi Q, Zhang J, Zhao X and Gu M: MiR-204-5p suppresses cell
proliferation by inhibiting IGFBP5 in papillary thyroid carcinoma.
Biochem Biophys Res Commun. 457:621–626. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Li W, Jin X, Zhang Q, Zhang G, Deng X and
Ma L: Decreased expression of miR-204 is associated with poor
prognosis in patients with breast cancer. Int J Clin Exp Pathol.
7:3287–3292. 2014.PubMed/NCBI
|
|
38
|
Mao J, Zhang M, Zhong M, Zhang Y and Lv K:
MicroRNA-204, a direct negative regulator of ezrin gene expression,
inhibits glioma cell migration and invasion. Mol Cell Biochem.
396:117–128. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Wu D, Niu X, Pan H, Zhou Y, Qu P and Zhou
J: MicroRNA-335 is downregulated in bladder cancer and inhibits
cell growth, migration and invasion via targeting ROCK1. Mol Med
Rep. 13:4379–4385. 2016.PubMed/NCBI
|
|
40
|
Bindels S, Mestdagt M, Vandewalle C,
Jacobs N, Volders L, Noël A, van Roy F, Berx G, Foidart JM and
Gilles C: Regulation of vimentin by SIP1 in human epithelial breast
tumor cells. Oncogene. 25:4975–4985. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Kurashige J, Kamohara H, Watanabe M,
Hiyoshi Y, Iwatsuki M, Tanaka Y, Kinoshita K, Saito S, Baba Y and
Baba H: MicroRNA-200b regulates cell proliferation, invasion, and
migration by directly targeting ZEB2 in gastric carcinoma. Ann Surg
Oncol. 19:(Suppl 3). S656–S664. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Chu PY, Hu FW, Yu CC, Tsai LL, Yu CH, Wu
BC, Chen YW, Huang PI and Lo WL: Epithelial-mesenchymal transition
transcription factor ZEB1/ZEB2 co-expression predicts poor
prognosis and maintains tumor-initiating properties in head and
neck cancer. Oral Oncol. 49:34–41. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Qi S, Song Y, Peng Y, Wang H, Long H, Yu
X, Li Z, Fang L, Wu A, Luo W, et al: ZEB2 mediates multiple
pathways regulating cell proliferation, migration, invasion, and
apoptosis in glioma. PLoS One. 7:e388422012. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Cai MY, Luo RZ, Chen JW, Pei XQ, Lu JB,
Hou JH and Yun JP: Overexpression of ZEB2 in peritumoral liver
tissue correlates with favorable survival after curative resection
of hepatocellular carcinoma. PLoS One. 7:e328382012. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Wu Q, Guo R, Lin M, Zhou B and Wang Y:
MicroRNA-200a inhibits CD133/1+ ovarian cancer stem cells migration
and invasion by targeting E-cadherin repressor ZEB2. Gynecol Oncol.
122:149–154. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Gemmill RM, Roche J, Potiron VA, Nasarre
P, Mitas M, Coldren CD, Helfrich BA, Garrett-Mayer E, Bunn PA and
Drabkin HA: ZEB1-responsive genes in non-small cell lung cancer.
Cancer Lett. 300:66–78. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Yang Z, Sun B, Li Y, Zhao X, Zhao X, Gu Q,
An J, Dong X, Liu F and Wang Y: ZEB2 promotes vasculogenic mimicry
by TGF-β1 induced epithelial-to-mesenchymal transition in
hepatocellular carcinoma. Exp Mol Pathol. 98:352–359. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Du Y, Wang L, Wu H, Zhang Y, Wang K and Wu
D: MicroRNA-141 inhibits migration of gastric cancer by targeting
zinc finger E-box-binding homeobox 2. Mol Med Rep. 12:3416–3422.
2015.PubMed/NCBI
|
|
49
|
Zhou J, Xie M, Shi Y, Luo B, Gong G, Li J,
Wang J, Zhao W, Zi Y, Wu X and Wen J: MicroRNA-153 functions as a
tumor suppressor by targeting SET7 and ZEB2 in ovarian cancer
cells. Oncol Rep. 34:111–120. 2015.PubMed/NCBI
|
|
50
|
Chen Z, Tang ZY, He Y, Liu LF, Li DJ and
Chen X: miRNA-205 is a candidate tumor suppressor that targets ZEB2
in renal cell carcinoma. Oncol Res Treat. 37:658–664. 2014.
View Article : Google Scholar : PubMed/NCBI
|