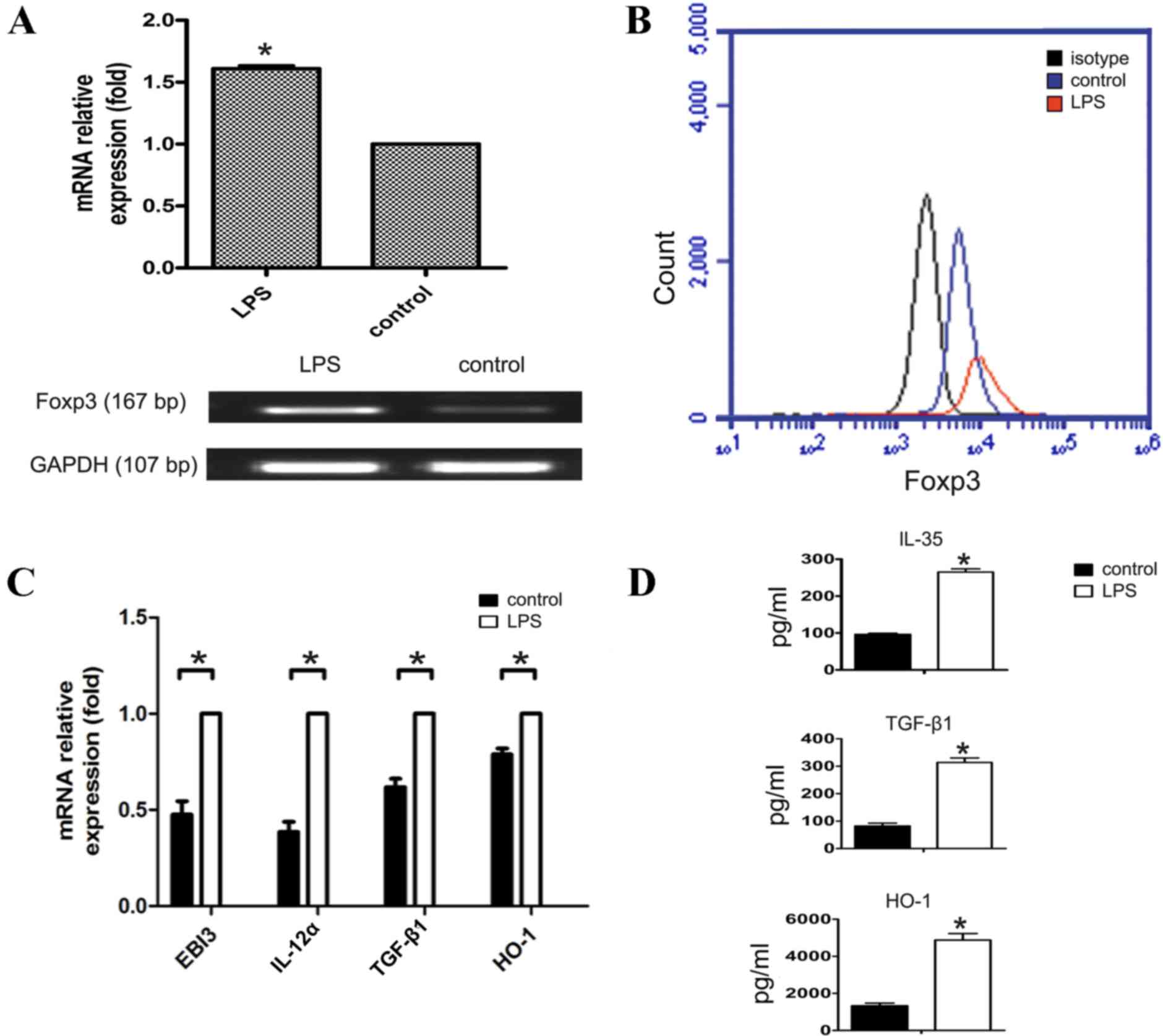
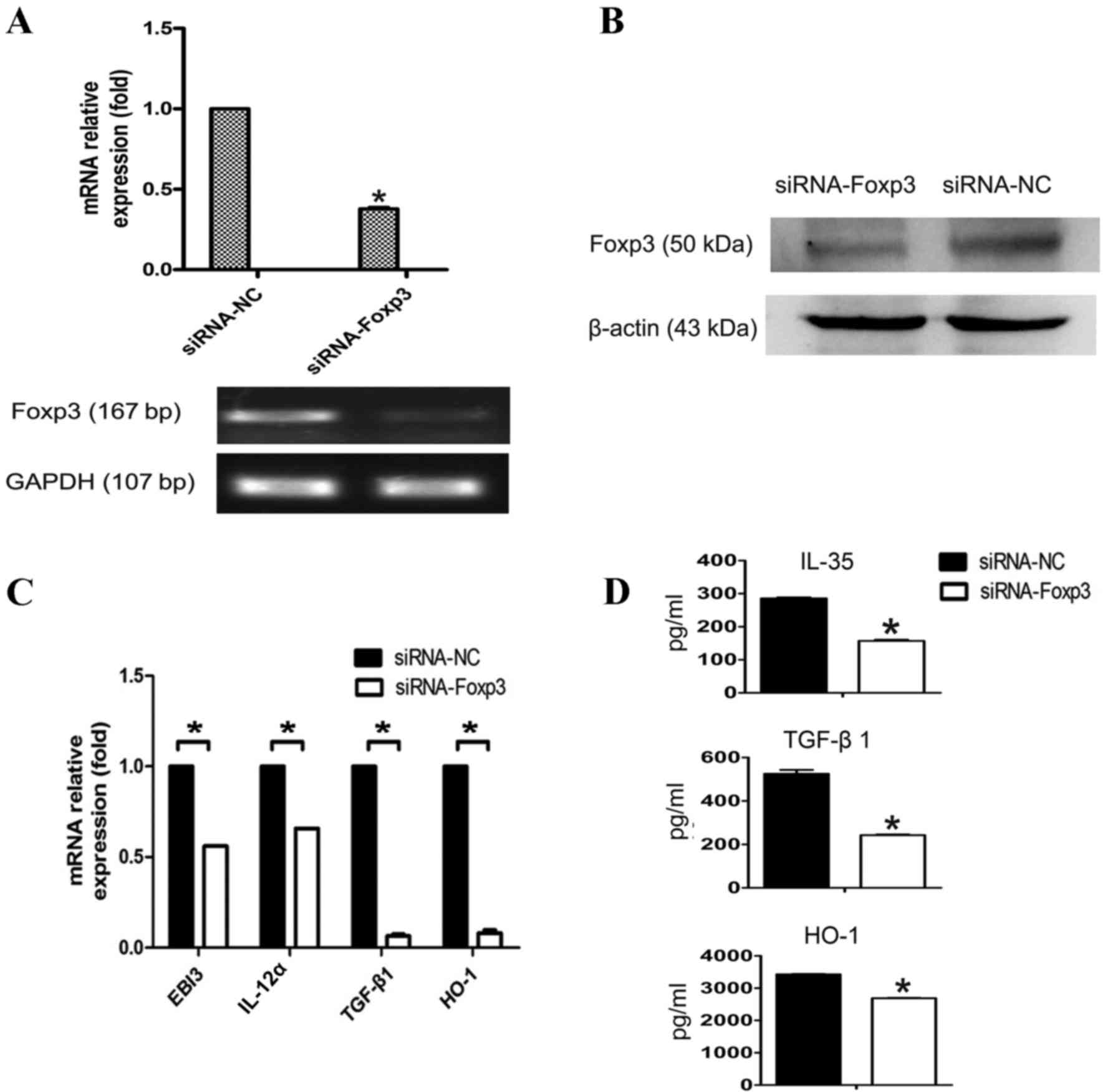
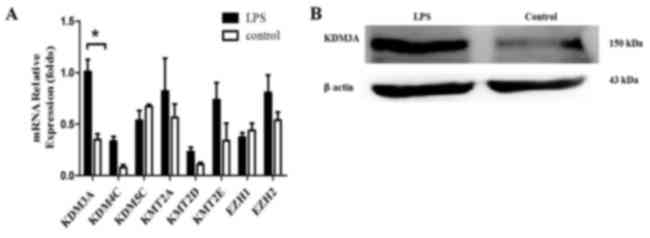
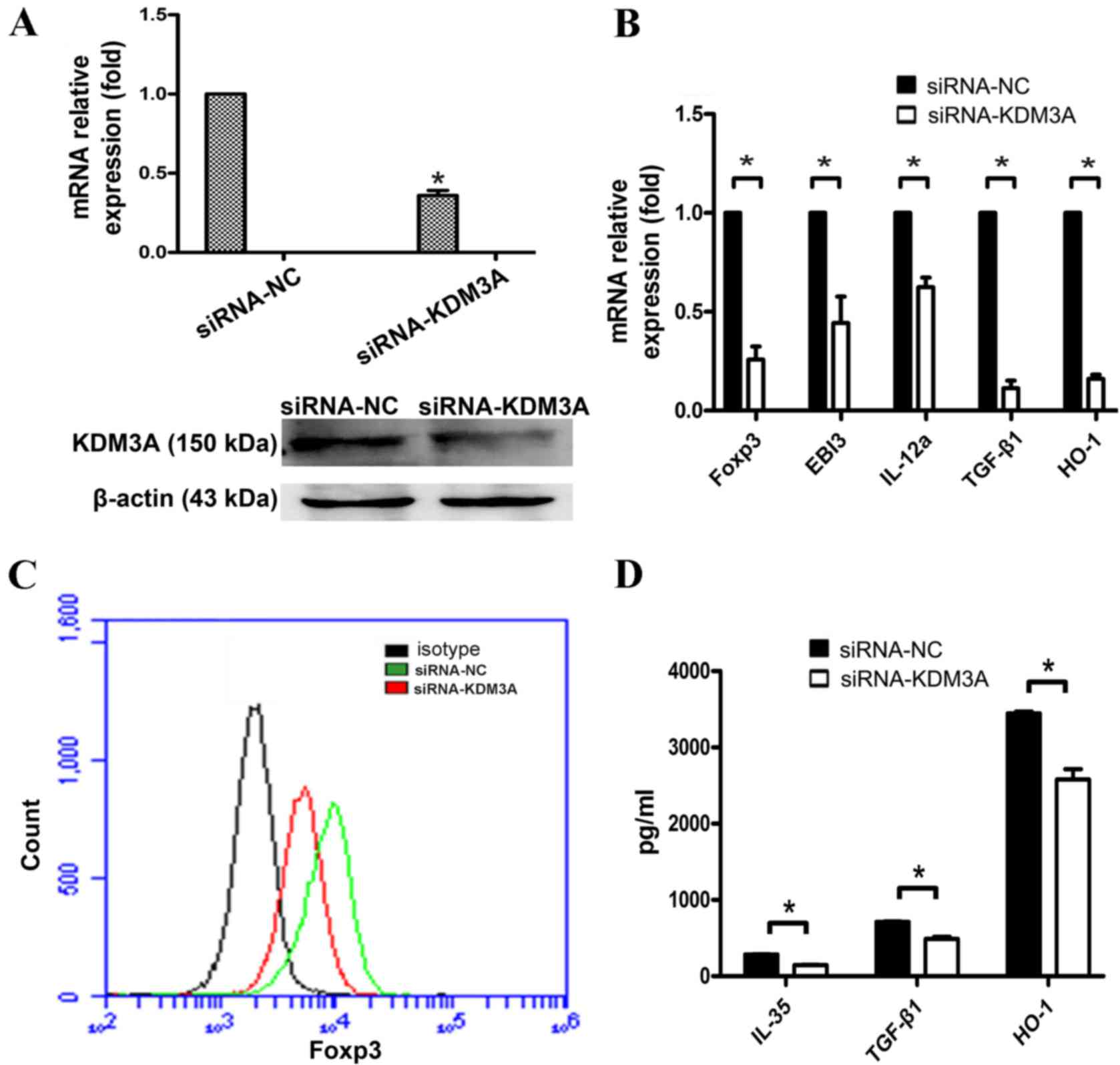
|
1
|
Alam H, Gu B and Lee MG: Histone
methylation modifiers in cellular signaling pathways. Cell Mol Life
Sci. 72:4577–4592. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Herzog M, Josseaux E, Dedeurwaerder S,
Calonne E, Volkmar M and Fuks F: The histone demethylase Kdm3a is
essential to progression through differentiation. Nucleic Acids
Research. 40:7219–7232. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Rea S, Eisenhaber F, OCarroll D, Strahl
BD, Sun ZW, Schmid M, Opravil S, Mechtler K, Ponting CP, Allis CD
and Jenuwein T: Regulation of chormatin structure by site-specific
histone H3 methyltransferase. Nature. 406:593–599. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Gol IMG and Bestor TH: Histone
modification and replacement in chromatin activation. Genes Dev.
16:1739–1742. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Suikki HE, Kujala PM, Tammela TL, van
Weerden WM, Vessella RL and Visakorpi T: Genetic alterations and
changes in expression of histone demethylases in prostate cancer.
Prostate. 70:889–898. 2010.PubMed/NCBI
|
|
6
|
Yang J, Ledaki I, Turley H, Gatter KC,
Montero JC, Li JL and Harris AL: Role of hypoxia-inducible factors
in epigenetic regulation via histone demethylases. Ann N Y Acad
Sci. 1177:185–197. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wang GG, Allis CD and Chi P: Chromatin
remodeling and cancer, part I: Covalent histone modification.
Trends Mol Med. 13:363–372. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Xiang Y, Zhu Z, Han G, Lin H, Xu L and
Chen CD: JMJD is a histone H3K27 demethylase. Cell Res. 17:850–857.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Nichol N, Dupéré-Richer D, Ezponda T,
Licht JD and Miller WH Jr: H3K27 methylation: A focal point of
epigenetic deregulation in cancer. Adv Cancer Res. 131:59–95. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yamane K, Toumazou C, Tsukada Y,
Erdjument-Bromage H, Tempst P, Wong J and Zhang Y: JHDM2A, a
Jmjc-containing H3K9 demethylase, facilitates transcription
activation by androgen receptor. Cell. 125:483–495. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Liu Y, Zhang Q, Ding Y, Li X, Zhao D, Zhao
K, Guo Z and Cao X: Histone lysine methyltransferase Ezh1 promotes
TLR-triggered inflammatory cytokines production by suppressing
tollip. J Immunol. 194:2838–2846. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Takeda K and Akira S: Toll-like receptors
in innate immunity. Int Immunol. 17:1–141. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Li TT, Ogiono S and Qian ZR: Toll-like
receptor signaling in colorectal cancer: Carcinogenesis to cancer
therapy. Word J Gastroenterol. 20:17699–17708. 2014.
|
|
14
|
Wang L, Zhao Y, Qian J, Sun L, Lu Y, Li H,
Li Y, Yang J, Cai Z and Yi Q: Toll-like receptor-4 signaling in
mantle cell lymphoma: Effects on tumor growth and immune evasion.
Cancer. 119:782–791. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Schmausser B, Andrulis M, Endrich S,
Mȕller-Hermelink HK and Eck M: Toll-like receptors TLR4, TLR5 and
TLR9 on gastric carcinoma cells: An implication for interaction
with Helicobacter pylori. Int J Med Microbiol. 295:179–185. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhou M, McFarland-Mancini MM, Funk HM,
Husseinzadeh N, Mounajjed T and Drew AF: Toll-like receptor
expression in normal ovary and ovarian tumors. Cancer Immunol
Immunother. 58:1375–2385. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wang EL, Qian ZR, Nakasono M, Tanahashi T,
Yoshimoto K, Bando Y, Kudo E, Shimada M and Sano T: High expression
of Toll-like receptor 4/myeloid differentiation factor 88 signals
correlates with poor prognosis in colorectal cancer. Br J Cancer.
102:908–915. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Milkova L, Voelcker V, Forstreuter I, Sack
U, Andereqq U, Simon JC and Maier-Simon C: The NF-kappaB signaling
pathway is involved in the LPS/IL-2-induced upregulation of Foxp3
expression in human CD4+CD25high regulatory T cells. Exp Dermatol.
19:29–37. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Li B, Samanta A, Song X, Furuuchi K,
Iacono KT, Kennedy S, Katsumata M, Saouaf SJ and Greene MI: FOXP3
ensembles in T-cell regulation. Immunol Rev. 212:99–131. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Schmitt EG and Williams CB: Generation and
function of induced regulatory T cells. Front Immunol. 4:1522013.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Karanikas V, Speletas M, Zamanakou M,
Kalala F, Loules G, Kerenidi T, Barda AK, Gourgoulianis KI and
Germenis AE: Foxp3 expression in human cancer cells. J Transl Med.
6:192008. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
He W, Liu Q, Wang L, Chen W, Li N and Cao
X: TLR4 signaling promotes immune escape of human lung cancer cells
by inducing immunosuppressive cytokines and apoptosis resistance.
Mol Immunol. 44:2850–2859. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Hinz S, Pagerols-Raluy L, Oberg HH,
Ammerpohl O, Grȕssel S, Sipos B, Grȕtzmann R, Pilarsky C,
Unqefroren H, Saeqer HD, et al: Foxp3 expression in pancreatic
carcinoma cells as a novel mechanism of immune evasion in cancer.
Cancer Res. 67:8344–8350. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Merlo A, Casalini P, Carcangiu ML,
Malventano C, Triulzi T, Mènard S, Tagliabue E and Balsari A: FOXP3
expression and overall survival in breast cancer. J Clin Oncol.
27:1746–1752. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wang L, Liu R, Li W, Chen C, Katoh H, Chen
GY, McNally B, Lin L, Zhou P, Zuo T, et al: Somatic single hits
inactivate the X-linked tumor suppressor FOXP3 in the prostate.
Cancer Cell. 16:336–346. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Triulzi T, Tagliabue E, Balsari A and
Casalini P: FOXP3 expression in tumor cells and implications for
cancer progression. J Cell Physiol. 228:30–35. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Fu HY, Li C, Yang W, Gai XD, Jia T, Lei YM
and Li Y: FOXP3 and TLR4 protein expression are correlated in
non-small cell lung cancer: Implications for tumor progression and
escape. Acta Histochem. 115:151–157. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Jia T, Fu H, Sun J, Zhang Y, Yang W and Li
Y: Foxp3 expression in A549 cells is regulated by Toll-like
receptor 4 through nuclear factor-κB. Mol Med Rep. 6:167–172.
2012.PubMed/NCBI
|
|
29
|
Κawai T and Akira S: The role of pattern
recognition receptors in innate immunity: Update on Toll like
receptors. Nat Immunol. 11:373–384. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Schmittgen TD and Livak KJ: Analyzing
real-time PCR data by the comparative C(T) method. Nat Protoc.
3:1101–1108. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Kim KH, Jo MS, Suh DS, Yoon MS, Shin DH,
Lee JH and Choi KU: Expression and signification of the TLR4/MyD88
signaling pathway in ovarian epithelial cancers. Word J Surg Oncol.
10:1932012. View Article : Google Scholar
|
|
32
|
Huang B, Zhao J, Li H, He KL, Chen Y, Chen
SH, Mayer L, Unkeless JC and Xiong H: Toll-like receptors on tumor
cells facilitate evasion of immune surveillance. Cancer Res.
65:5009–5014. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Wang L, Zhao Y, Qian J, Sun L, Lu Y, Li H,
Li Y, Yanq J, Cai Z and Yi Q: Toll-like receptor-4 signaling in
mantle cell lymphoma: Effects on tumor growth and immune evasion.
Cancer. 119:782–791. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Goto Y, Arigami T, Kitago M, Nquyen SL,
Narita N, Ferrone S, Morton DL, Irie RF and Hoon DS: Activation of
Toll-like receptors 2, 3 and 4 on human melanoma cells induces
inflammatory factors. Mol Cancer Ther. 7:3642–3653. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Hori S, Nomura T and Sakaguchi S: Control
of regulatory T cell development by the transcription factor Foxp3.
Science. 299:1057–1061. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Laouar Y, Welte T, Fu XY and Flavell RA:
STAT3 is required for Flt3L-dependent dendritic cell
differentiation. Immunity. 19:903–912. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Welte T, Koch F, Schuler G, Lechner J,
Doppler W and Heufler C: Granulocyte-macrophage colony-stimulating
factor induces a unique set of STAT factors in murine dendritic
cells. Eur J Immunol. 27:2737–2740. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Tsukada Y, Fang J, Erdjument-Bromage H,
Warren ME, Borchers CH, Tempst P and Zhang Y: Histone demethylation
by a family of Jmjc domain-containing protein. Nature. 439:811–816.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Okada Y, Scott G, Ray MK, Mishina Y and
Zhang Y: Histone demethylase JHDM2A is critical for Tnp1 and Prm1
transcription and spermatogenesis. Nature. 450:119–123. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Liu Z, Zhou S, Liao L, Chen X, Meisrrich M
and Xu J: Jmjd1a demethylase-regulated histone modification is
essential for cAMP-response element modulator-regulated gene
expression and spermatogenesis. J Biol Chem. 285:2758–2770. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Parrish JK, Sechler M, Winn RA and Jedicka
P: The histone demethylase KDM3A is a microRNA-22-regulated tumor
promoter in ewing sarcoma. Oncogene. 34:257–262. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Uemura M, Yamamoto H, Takemasa I, Mimori
K, Hemmi H, Mizushima T, Ikeda M, Sekimoto M, Matsuura N, Doki Y
and Mori M: Jumonji domain containing 1A is a novel prognostic
marker for colorectal cancer: In vivo identification from hypoxic
tumor cells. Clin Cancer Res. 16:4636–4646. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Mahajan K, Lawrence HR, Lawrence NJ and
Mahajan NP: ACK1 tyrosine kinase interacts with histone demethylase
KDM3A to regulate the mammary tumor oncogene HOXA1. J Biol Chem.
289:28179–28191. 2014. View Article : Google Scholar : PubMed/NCBI
|