Introduction
Angiosarcoma is a rare malignant vascular tumor and
is difficult to diagnose and treat (1). It may be characterized by rapidly
proliferating and extensively infiltrating anaplastic cells, which
are derived from blood vessels, and lining irregular blood-filled
spaces (2,3). Angiosarcoma is derived from mesenchymal
cells and usually originates from the liver, breast, spleen, bone
or heart (2,4–6).
Angiosarcoma accounts for between 1 and 2% of all sarcomas, and its
overall 5-year survival rate is <20%, owing to the high
recurrence and distant metastasis rates (2). In addition, metastasis usually occurs in
the liver, lung, bone and lymph nodes (7). Treatment of angiosarcoma is multifaceted
and primarily consists of radiotherapy, surgery and chemotherapy
(8).
Propranolol is a non-selective β-blocker and may
inhibit the growth of angiosarcoma by affecting the proliferation
and differentiation of angiosarcoma tumor cells, thus being
considered a promising treatment to delay surgery (9–11).
However, its underlying molecular mechanisms and pharmacodynamics
of the effects on angiosarcoma remain obscure, and the potential
target genes involved in the proliferation and differentiation
processes of angiosarcoma tumor cells also require
investigation.
Gene microarray is widely used as an effective
technology to detect the gene expression in cells and tissues at
different disease stages of cancer. Thus, it may aid in the
identification of novel signaling pathways or molecular mechanisms
associated with tumorigenesis.
In the present study, the differentially expressed
genes (DEGs) in angiosarcoma tumor cells treated with propranolol
compared with the control group were identified via a
bioinformatics-based method. Furthermore, enenrichment analysis,
protein-protein interaction (PPI) network construction and module
analysis were performed. These analyses aided in the identification
of essential genes associated with angiosarcoma, such as AXL
receptor tyrosine kinase (AXL), coatomer subunit α, DR1-associated
protein 1, ERBB receptor feedback inhibitor 1, family with sequence
similarity 195 member A, expressed sequence AA467197,
apoptosis-associated tyrosine kinase, ATP-binding cassette
subfamily A member 7, acyl-CoA dehydrogenase family member 9 and
acyl-CoA-binding domain containing 6. Thus, this may contribute to
understanding the molecular mechanism underlying angiosarcoma in
order to identify potential gene targets for the diagnosis and
treatment of patients with angiosarcoma.
Materials and methods
mRNA expression microarray data
The standardized mRNA expression profile GSE42534
(9) was downloaded from the Gene
Expression Omnibus (www.ncbi.nlm.nih.gov/geo/) database, including 3
samples without propranolol treatment (the control group), 3
samples with propranolol treatment for 4 h and 3 samples with
propranolol treatment for 24 h.
Identification and grouping of
differentially expressed genes
The DEGs in angiosarcoma tumor cells of the
propranolol treatment groups compared with the control group were
obtained using the limma package of R (http://bioconductor.org/packages/release/bioc/html/limma.html)
(12). They were designated DEGs-4 h
and DEGs-24 h. The DEGs-24 h were divided into 2 sets. Set 1
contained those DEGs also contained in the DEGs-4 h group. Set 2
contained the remainder of the DEGs. For the sake of accuracy, all
DEGs were identified according to the following criteria:
P<0.001;|log2 (fold-change) |≥1.
Gene ontology (GO) and pathway
enrichment analysis
In order to explore the potential biological
processes that were altered, GO and Kyoto Encyclopedia of Genes and
Genomes (KEGG) pathway enrichment analysis were performed using the
Database for Annotation, Visualization and Integrated Discovery
(DAVID; david.abcc.ncifcrf.gov/) (13). The GO terms and the KEGG pathways were
identified with the criterion P<0.05.
Construction of protein-protein
interaction (PPI) networks
The two PPI networks for sets 1 and 2 were
constructed using the Search Tool for the Retrieval of Interacting
Genes/Proteins (STRING) (14)
database, termed PPI 1 and PPI 2, respectively, and visualized
using Cytoscape software (version 3.4.0; http://www.cytoscape.org/) (15). STRING, which manipulates the
interactions between genes or proteins from multiple sources, was
used to identify the interactions of DEGs. A combined score (a
representation of reliability of interactions) >0.4 was used as
the threshold for the selection of interaction pairs. Modules of
the two PPI networks were analyzed using the Multi Contrast Delayed
Enhancement plug-in of Cytoscape (16). When the combined score was >1.5,
function enrichment analysis of all enrolled DEGs was performed
using DAVID, and the GO terms and the KEGG pathways with P<0.05
were identified. Topological structures of the two PPI networks
were analyzed using tYNA (tyna.gersteinlab.org/tyna) (17), and potential target genes, whereby the
degree of node attributes was ≥10, were identified. Degree
represents the number of direct interactions a node has with with
other nodes.
Results
Identification of DEGs
A total of 543 DEGs (242 up- and 301 downregulated)
and 2,025 DEGs (1,107 up- and 918 downregulated) were identified in
angiosarcoma tumor cells treated with propranolol (DEGs-4 h and
DEGs-24 h, respectively) compared with the control group. A total
of 401 DEGs (set 1) were involved in DEGs-4 h and DEGs-24 h,
including metallothionein 1, heme oxygenase 1, WW domain-binding
protein 2 and sequestosome 1. Among set 1, 179 DEGs in the DEGs-4 h
group were upregulated, of which 170 DEGs were upregulated and 9
DEGs (2410011G03Rik, 2810417H13Rik, ATPase inhibitory factor 1, G2
and S-phase expressed 1, LSM5 homolog U6 small nuclear RNA and mRNA
degradation associated, non-SMS condensing I complex subunit H,
Rp127, ubiquitin-40A ribosomal protein S27a precursor and zinc
finger CCHC-type-containing 8) were downregulated in the DEGs-24 h
group. Similarly, 222 DEGs of the DEGs-4 h group were
downregulated, of which 196 DEGs were downregulated and 26 DEGs
[D730049H07Rik, desert hedgehog, dual-specificity phosphatase 7,
endothelin 1, ETS proto-oncogene 1, general receptor for
phosphoinositides, mitogen-associated protein kinase 6, midnolin,
myeloid-associated differentiation marker, lysophosphatidic acid
receptor 6, platelet-derived growth factor subunit A, PDZ and LIM
domain (Pdlim) 1, Pdlim7, plexin A2, phosphatidic acid phosphatase
type 2B, Ppmlf, regulator of G-protein signaling 16, ras homolog
family member B, roundabout guidance receptor 4, sterile α motif
domain-containing 4, solute carrier family 2 member 1, solute
carrier family 9 isoform A3 regulatory factor 2, tissue inhibitor
of metalloproteinase 3, tumor necrosis factor-α-induced protein 2,
trophoblast glycoprotein and WNT1-inducible signaling pathway
protein 1] were upregulated in the DEGs-24 h group.
Functional and pathway enrichment
analysis of sets 1 and 2
The top 20 most significantly enriched GO terms of
sets 1 and 2 are presented in Table IA
and B, respectively. Among them, 28 terms were coincident
(Table IC). The enriched KEGG
pathways of sets 1 and 2 are presented in Table IIA and B, respectively.
 | Table I.Significantly enriched and coincident
GO terms in sets 1 and 2. |
Table I.
Significantly enriched and coincident
GO terms in sets 1 and 2.
| A, Top 20 most
significantly enriched GO terms in set 1 |
|---|
|
|---|
| GO ID | GO name | Gene number | P-value |
|---|
| GO:0005730 | Nucleolus | 23 | 0.000000006 |
| GO:0016126 | Sterol biosynthetic
process | 8 | 0.000000580 |
| GO:0031974 | Membrane-enclosed
lumen | 43 | 0.000000604 |
| GO:0001525 | Angiogenesis | 12 | 0.000017900 |
| GO:0070013 | Intracellular
organelle lumen | 38 | 0.000025500 |
| GO:0043233 | Organelle lumen | 38 | 0.000027000 |
| GO:0006694 | Steroid biosynthetic
process | 9 | 0.000027800 |
| GO:0006695 | Cholesterol
biosynthetic process | 6 | 0.000037100 |
| GO:0048514 | Blood vessel
morphogenesis | 14 | 0.000037200 |
| GO:0016125 | Sterol metabolic
process | 9 | 0.000050400 |
| GO:0001568 | Blood vessel
development | 15 | 0.000081700 |
| GO:0031981 | Nuclear lumen | 31 | 0.000082000 |
| GO:0001944 | Vasculature
development | 15 | 0.000106000 |
| GO:0005773 | Vacuole | 13 | 0.000118000 |
| GO:0008610 | Lipid biosynthetic
process | 16 | 0.000120000 |
| GO:0005764 | Lysosome | 12 | 0.000148000 |
| GO:0000323 | Lytic vacuole | 12 | 0.000156000 |
| GO:0043232 | Intracellular
non-membrane-bounded organelle | 51 | 0.000304000 |
| GO:0043228 |
Non-membrane-bounded organelle | 51 | 0.000304000 |
| GO:0042127 | Regulation of cell
proliferation | 22 | 0.000387000 |
|
| B, Top 20 most
significantly enriched GO terms in set 2 |
|
| GO ID | GO name | Gene number | P-value |
|
| GO:0030529 | Ribonucleoprotein
complex | 100 | 0.000000000000 |
| GO:0005739 | Mitochondrion | 178 | 0.000000000000 |
| GO:0005840 | Ribosome | 52 | 0.000000000000 |
| GO:0044429 | Mitochondrial
part | 84 | 0.000000000001 |
| GO:0003735 | Structural
constituent of ribosome | 39 | 0.000000000002 |
| GO:0043233 | Organelle
lumen | 142 | 0.000000000003 |
| GO:0031974 | Membrane-enclosed
lumen | 145 | 0.000000000005 |
| GO:0070013 | Intracellular
organelle lumen | 141 | 0.000000000006 |
| GO:0043228 |
Non-membrane-bounded organelle | 208 | 0.000000000014 |
| GO:0043232 | Intracellular
non-membrane-bounded organelle | 208 | 0.000000000014 |
| GO:0006412 | Translation | 58 | 0.000000000047 |
| GO:0031090 | Organelle
membrane | 108 | 0.000000000050 |
| GO:0005681 | Spliceosome | 33 | 0.000000000056 |
| GO:0006396 | RNA processing | 70 | 0.000000000140 |
| GO:0008380 | RNA splicing | 42 | 0.000000000438 |
| GO:0031967 | Organelle
envelope | 79 | 0.000000000456 |
| GO:0031975 | Envelope | 79 | 0.000000000543 |
| GO:0019866 | Organelle inner
membrane | 54 | 0.000000001140 |
| GO:0006397 | mRNA
processing | 48 | 0.000000002120 |
| GO:0016071 | mRNA metabolic
process | 51 | 0.000000010600 |
|
| C, Coincident
enriched GO terms in sets 1 and 2 |
|
| GO ID | GO name | GO ID | GO name |
|
| GO:0000166 | Nucleotide
binding | GO:0031981 | Nuclear lumen |
| GO:0005730 | Nucleolus | GO:0032553 | Ribonucleotide
binding |
| GO:0005773 | Vacuole | GO:0032555 | Purine
ribonucleotide binding |
| GO:0005783 | Endoplasmic
reticulum | GO:0034404 | Nucleobase,
nucleoside and nucleotide biosynthetic process |
| GO:0005829 | Cytosol | GO:0034654 | Nucleobase,
nucleoside, nucleotide and nucleic acid biosynthetic process |
| GO:0006334 | Nucleosome
assembly | GO:0034728 | Nucleosome
organization |
| GO:0006364 | rRNA
processing | GO:0042254 | Ribosome
biogenesis |
| GO:0006396 | RNA processing | GO:0043228 |
Non-membrane-bounded organelle |
| GO:0009165 | Nucleotide
biosynthetic process | GO:0043232 | Intracellular
non-membrane-bounded organelle |
| GO:0016072 | rRNA metabolic
process | GO:0043233 | Organelle
lumen |
| GO:0017076 | Purine nucleotide
binding | GO:0044271 | Nitrogen compound
biosynthetic process |
| GO:0022613 | Ribonucleoprotein
complex biogenesis | GO:0046907 | Intracellular
transport |
| GO:0030529 | Ribonucleoprotein
complex | GO:0051726 | Regulation of cell
cycle |
| GO:0031974 | Membrane-enclosed
lumen | GO:0070013 | Intracellular
organelle lumen |
 | Table II.Enriched KEGG pathways in sets 1 and
2. |
Table II.
Enriched KEGG pathways in sets 1 and
2.
| A, Enriched KEGG
pathways in set 1 |
|---|
|
|---|
| Term | Count | P-value |
|---|
| mmu04115: p53
signaling pathway | 9 | 0.000105 |
| mmu00100: Steroid
biosynthesis | 5 | 0.000398 |
| mmu00900: Terpenoid
backbone biosynthesis | 4 | 0.003012 |
| mmu04142:
Lysosome | 9 | 0.004013 |
| mmu00600:
Sphingolipid metabolism | 5 | 0.012340 |
| mmu00240:
Pyrimidine metabolism | 7 | 0.017197 |
| mmu00270: Cysteine
and methionine metabolism | 4 | 0.033547 |
| mmu00650: Butanoate
metabolism | 4 | 0.044913 |
| mmu05214:
Glioma | 5 | 0.048883 |
|
| B, Enriched KEGG
pathways in set 2 |
|
| Term | Count | P-value |
|
| mmu03040:
Spliceosome | 37 | 0.000000 |
| mmu00190: Oxidative
phosphorylation | 31 | 0.000001 |
| mmu03010:
Ribosome | 22 | 0.000026 |
| mmu04142:
Lysosome | 24 | 0.000291 |
| mmu05211: Renal
cell carcinoma | 17 | 0.000350 |
| mmu00480:
Glutathione metabolism | 13 | 0.001727 |
| mmu05016:
Huntington's disease | 28 | 0.006229 |
| mmu05012:
Parkinson's disease | 22 | 0.007000 |
| mmu05222: Small
cell lung cancer | 16 | 0.007770 |
| mmu03030: DNA
replication | 9 | 0.010537 |
| mmu04666: Fc gamma
R-mediated phagocytosis | 17 | 0.012796 |
| mmu04114: Oocyte
meiosis | 19 | 0.013122 |
| mmu03018: RNA
degradation | 12 | 0.016095 |
| mmu04110: Cell
cycle | 20 | 0.018766 |
| mmu05200: Pathways
in cancer | 41 | 0.020303 |
| mmu04662: B cell
receptor signaling pathway | 14 | 0.024365 |
| mmu05212:
Pancreatic cancer | 13 | 0.024948 |
| mmu00600:
Sphingolipid metabolism | 9 | 0.030444 |
| mmu00980:
Metabolism of xenobiotics by cytochrome P450 | 12 | 0.031062 |
| mmu00330: Arginine
and proline metabolism | 10 | 0.043757 |
| mmu00860: Porphyrin
and chlorophyll metabolism | 7 | 0.046693 |
| mmu00511: Other
glycan degradation | 5 | 0.048603 |
| mmu04062: Chemokine
signaling pathway | 24 | 0.056122 |
| mmu05010:
Alzheimer's disease | 24 | 0.056122 |
| mmu05215: Prostate
cancer | 14 | 0.056323 |
| mmu04620: Toll-like
receptor signaling pathway | 15 | 0.056726 |
| mmu03410: Base
excision repair | 8 | 0.061961 |
| mmu00230: Purine
metabolism | 21 | 0.066837 |
| mmu00982: Drug
metabolism | 12 | 0.068874 |
| mmu04920:
Adipocytokine signaling pathway | 11 | 0.073182 |
| mmu04810:
Regulation of actin cytoskeleton | 27 | 0.075019 |
Construction of the PPI networks for
sets 1 and 2 and analysis of modules
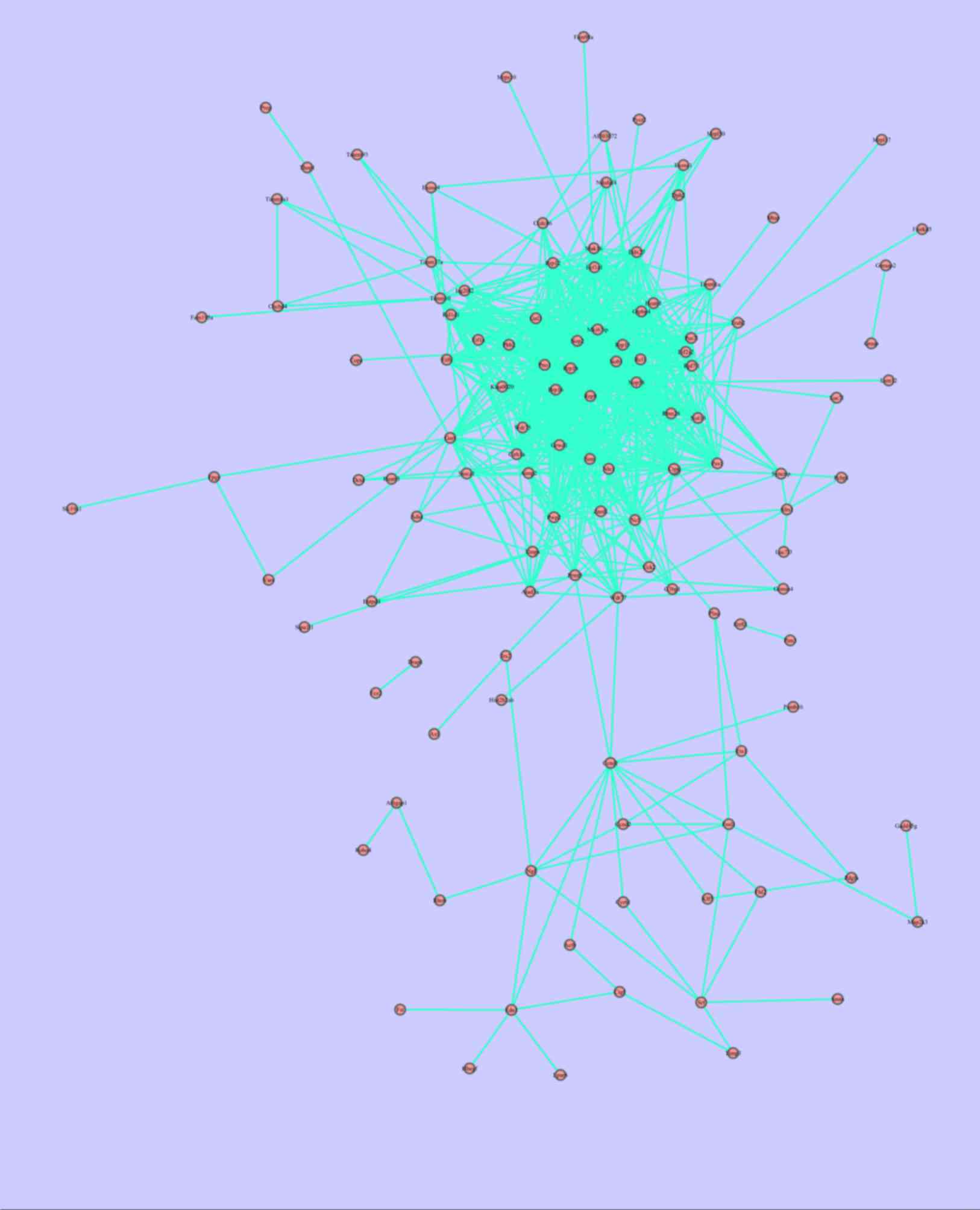
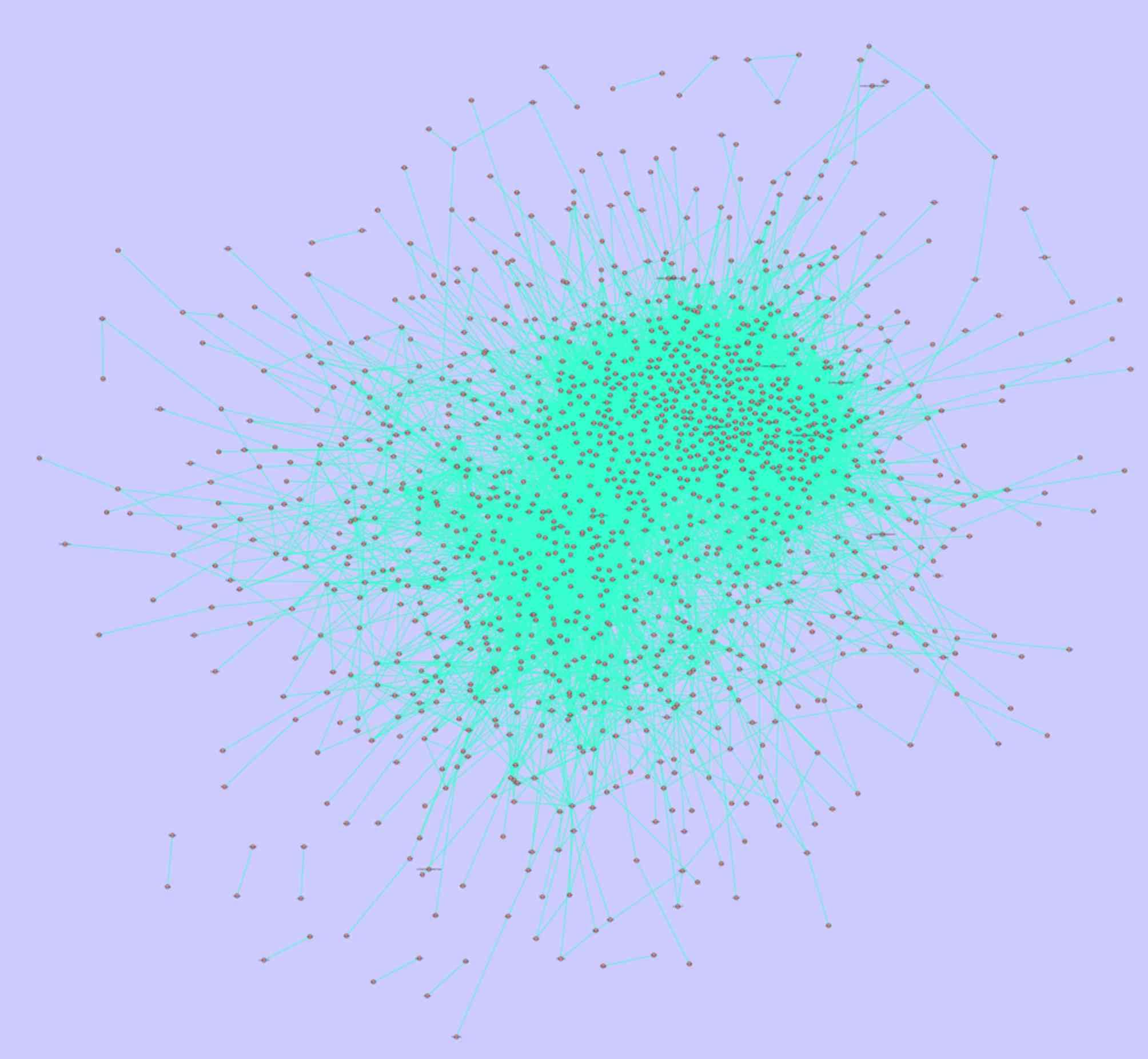
The PPI networks of PPI 1 and PPI 2 are presented in
Figs. 1 and 2. A total of 121 nodes and 700 associated
pairs were involved in PPI 1, whereas 1,324 nodes and 11,839
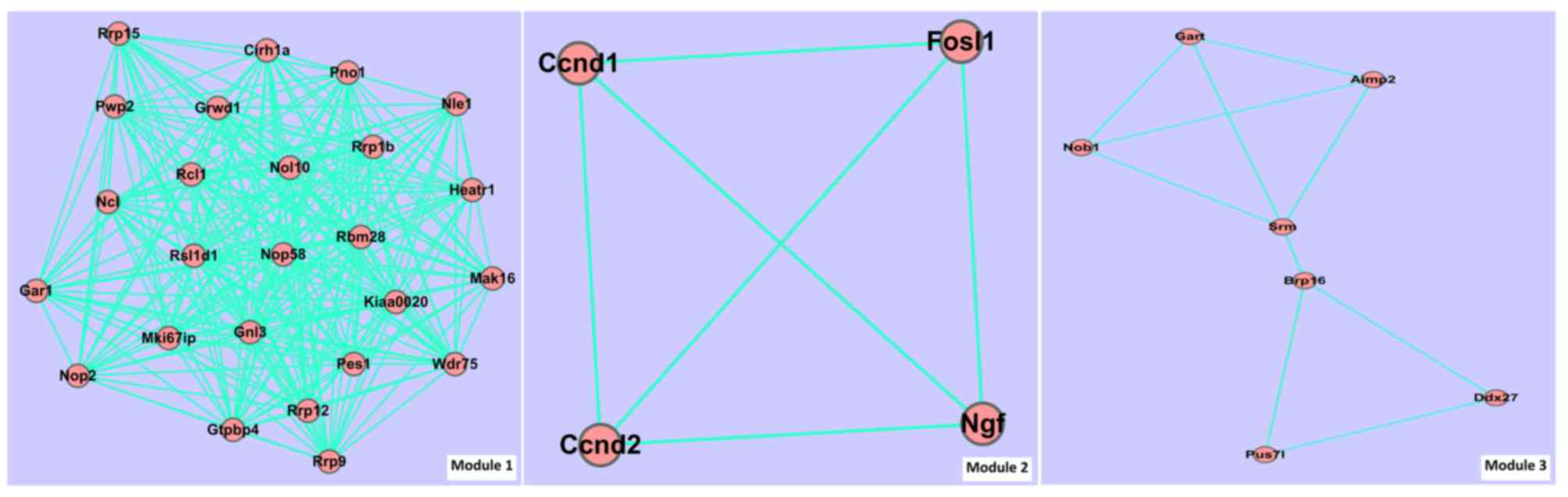
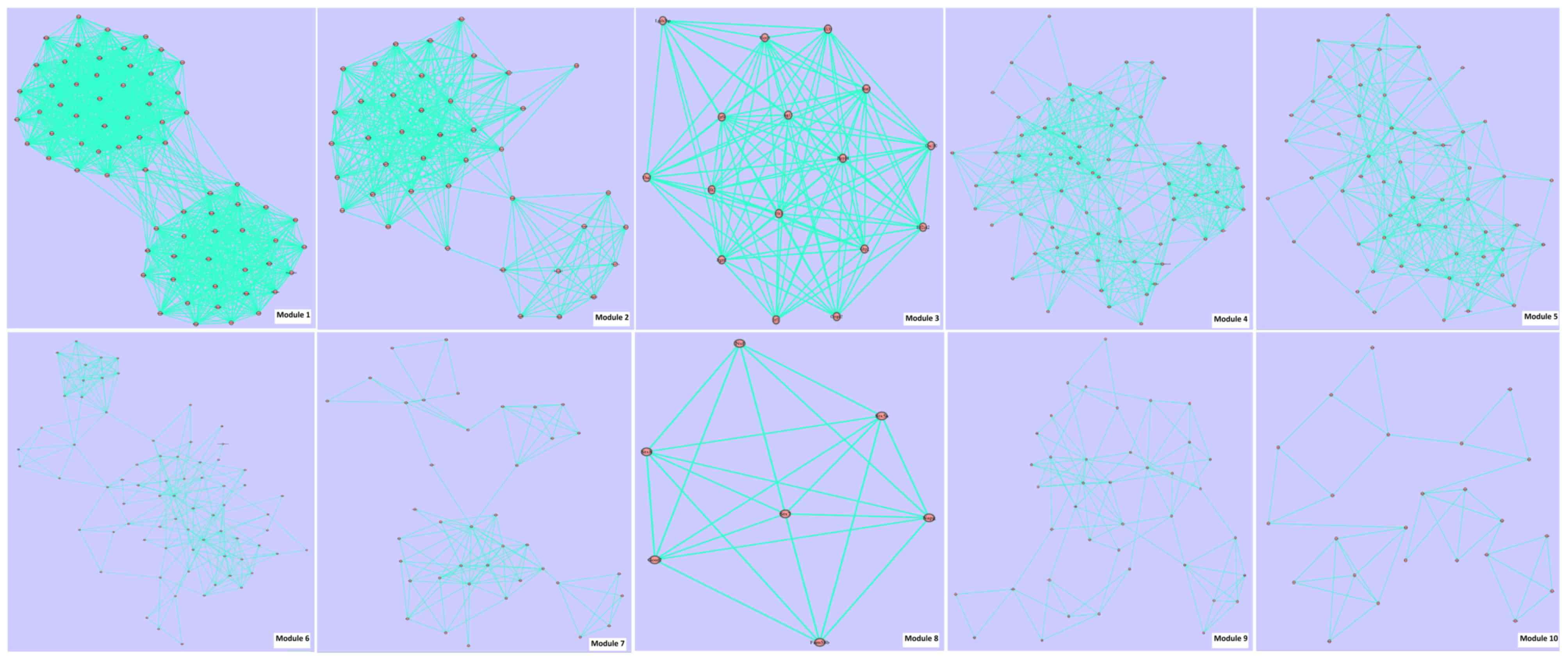
associated pairs were involved in PPI 2. Fig. 3 and Table
IIIA present the module information of PPI 1. Fig. 4 and Table
IIIB present the module information of PPI 2.
 | Table III.Module information of the
protein-protein interaction networks for sets 1 and 2. |
Table III.
Module information of the
protein-protein interaction networks for sets 1 and 2.
| A, Module
information of the protein-protein interaction network of set
1 |
|---|
|
|---|
| Module ID | Score | Gene number | Edge number |
|---|
| 1 | 10.4 | 25 | 260 |
| 2 |
1.5 | 4 |
6 |
| 3 |
1.5 | 7 | 10 |
|
| B, Module
information of the protein-protein interaction network of set
2 |
|
| Module ID | Score | Gene number | Edge number |
|
| 1 | 17.1 | 70 | 1195 |
| 2 |
9.4 | 41 |
387 |
| 3 |
6.8 | 16 |
109 |
| 4 |
6.4 | 70 |
446 |
| 5 |
5.2 | 56 |
292 |
| 6 |
3.6 | 74 |
267 |
| 7 |
2.9 | 39 |
113 |
| 8 |
2.9 | 7 |
20 |
| 9 |
2.5 | 40 |
99 |
| 10 |
1.7 | 24 |
41 |
Extent of enriched function and
topological structure analysis of the PPI networks
There were 20 GO terms (including nucleolus,
intracellular organelle lumen, membrane-enclosed lumen, ribosome
biogenesis and RNA processing) and no KEGG pathways enriched in
module 1 of PPI 1. The numbers of the enriched module functions of
PPI 2 are presented in Table IV. The
results identified that no enriched KEGG pathways appeared in
modules 1 and 9 of PPI 2. A total of 45 and 593 potential target
genes were obtained according to the node degrees of PPI 1 and PPI
2, and the top 10 nodes (potential target genes) which were
associated with the other nodes in the PPI networks for sets 1 and
2 are presented in Table VA and
VB, respectively.
 | Table IV.Enriched function numbers of modules
of the protein-protein interaction network of set 2. |
Table IV.
Enriched function numbers of modules
of the protein-protein interaction network of set 2.
| Modules | Enriched GO term
numbers | Enriched KEGG
pathway number |
|---|
| Module 1 |
0 | 0 |
| Module 2 | 47 | 1 |
| Module 3 | 10 | 1 |
| Module 4 | 122 | 6 |
| Module 5 | 62 | 5 |
| Module 6 | 90 | 11 |
| Module 7 | 105 | 15 |
| Module 8 | 12 | 1 |
| Module 9 | 154 | 0 |
| Module 10 | 22 | 6 |
 | Table V.Top 10 nodes most significantly
associated with other nodes in the protein-protein interaction
network of sets 1 and 2. |
Table V.
Top 10 nodes most significantly
associated with other nodes in the protein-protein interaction
network of sets 1 and 2.
| A, Top 10 nodes
most significantly associated with other nodes in the
protein-protein interaction network of set 1 |
|---|
|
|---|
| Gene symbol | Degree | Clustering
coefficient | Eccentricity | Betweenness
centrality |
|---|
| AXL | 45 | 0 | 6 | 0 |
| COPA | 44 | 0 | 7 | 0 |
| DRAP1 | 44 | 0 | 2 | 0 |
| ERRFI1 | 41 | 0 | 2 | 0 |
| FAM195A | 38 | 0 | 8 | 0 |
| FAM98A | 36 | 0 | 8 | 0 |
| FASTKD5 | 36 | 0 | 8 | 0 |
| FEZ2 | 33 | 0 | 2 | 0 |
| FST | 33 | 0 | 6 | 0 |
| GADD45G | 33 | 0 | 7 | 0 |
|
| B, Top 10 nodes
most significantly associated with other nodes in the
protein-protein interaction network of set 2 |
|
| Gene symbol | Degree | Clustering
coefficient | Eccentricity | Betweenness
centrality |
|
| AA467197 | 184 | 0 | 2 | 0 |
| AATK | 120 | 0 | 8 | 0 |
| ABCA7 | 120 | 0 | 7 | 0 |
| ACAD9 | 116 | 0 | 2 | 0 |
| ACBD6 | 113 | 0 | 8 | 0 |
| ACSL3 | 111 | 0 | 9 | 0 |
| AFG3L1 | 109 | 0 | 7 | 0 |
| AGAP1 | 109 | 0 | 8 | 0 |
| AHNAK | 106 | 0 | 9 | 0 |
| ANGEL2 | 106 | 0 | 7 | 0 |
Discussion
Numerous studies have demonstrated the selective
cytotoxicity and relative safety of propranolol on vascular tumors,
and laid the groundwork for the notable efficacy and the
suppressive ability of propranolol on angiosarcoma (9–11,18–20). In
the present study, it was found that the number of DEGs-24 h was
higher compared with the number of DEGs-4 h. In addition, nearly
all of the DEGs-4 h overlapped with and were contained in the
DEGs-24 h group. Furthermore, differential expression (upregulated
or downregulated) of DEGs-24 h was more evident compared with
DEGs-4 h. This indicated that the 401 overlapping DEGs in set 1
were important in the effects of propranolol on angiosarcoma tumor
cells. Notably, 9 upregulated DEGs of the DEGs-4 h group were
downregulated in the DEGs-24 h group, whereas 26 downregulated DEGs
of the DEGs-4 h group were upregulated in the DEGs-24 h group. It
was possible that these genes perform multiple roles in the effect
of propranolol on angiosarcoma; however, this conjecture requires
additional experimental verification.
The enriched GO terms of set 1 primarily contained
‘angiogenesis, blood vessel morphogenesis, vasculature
development’, ‘sterol biosynthetic process, cholesterol
biosynthetic process, lipid biosynthetic process’, ‘lysosome, lytic
vacuole, vacuole’, and ‘nucleolus, intracellular
non-membrane-bounded organelle, regulation of cell proliferation’.
It is well known that lipid metabolism may affect the development
of blood vessels (21–23) and various organelles involved in
various biological processes (23,24). Cell
proliferation is an essential process in the development of blood
vessels (25). According to Table IA, the majority of enriched GO terms
of set 1 were associated with the biological processes of blood
vessels, whereas the enriched GO terms of set 2 were primarily
associated with energy metabolism (including ribosome, structural
constituent of ribosome), protein transfer (including
ribonucleoprotein complex, ribosome, membrane-enclosed lumen) and
compounds biosynthesis (including RNA processing, mRNA metabolic
process and envelope). The overlapping enriched GO terms of sets 1
and 2 were primarily involved in nucleic acid metabolism,
nucleotide biosynthesis and nucleic acid binding. Therefore, it was
concluded that propranolol affected angiosarcoma primarily by
influencing the biological processes of blood vessels in the early
stage and by effecting the biological metabolism and transfer
processes in the later stage. The enriched KEGG pathways of set 1
were tumor-associated biological processes, including the p53
signaling pathway and cysteine and methionine metabolism. In the
later stage, the enriched KEGG pathways were more extensive,
including the ribosome signaling pathway, lysosome signaling
pathway, Huntington's disease and Parkinson's disease.
According to the topological structure analysis of
the PPI networks, certain potential biomarkers were identified,
including AXL, coatomer subunit α, DR1-associated protein 1, ERBB
receptor feedback inhibitor 1, family with sequence similarity 195
member A, expressed sequence AA467197, apoptosis-associated
tyrosine kinase, ATP-binding cassette subfamily A member 7,
acyl-CoA dehydrogenase family member 9 and acyl-CoA-binding domain
containing 6. According to Table VA,
AXL was the most significantly meaningful gene in the early
stage. AXL is a member of the tyrosine kinase receptor
family and is associated with cell adhesion and recognition, cell
proliferation, apoptosis, blood coagulation and inflammation
(26). It performs important roles in
the occurrence and development of various tumors, including the
inhibition of tumor cell apoptosis, the involvement in tumor
angiogenesis and cellular invasion (27–30).
Following its original identification, the upregulation of
Axl has been reported in a variety of hematopoietic tumors,
including leukemia and melanoma (31–35).
Furthermore, previous studies have demonstrated that Axl may
also perform a role in a number of chemotherapy-resistant cancers
(36,37). In the present study, it was proposed
that Axl may be a potential target in the early stage of
angiosarcoma treated with propranolol. This discovery may indicate
an important direction for future studies. Similarly,
AA467197 may be a potential biomarker in the late stage of
angiosarcoma treated with propranolol. It is a key point of the
effects of propranolol on angiosarcoma to identify and develop
small-molecule drugs with the potential to selectively inhibit
Axl and AA467197 expression and their signaling
pathways.
Acknowledgements
The present study was supported by the Municipal
Science and Technology Commission of Tianjin (grant no.
15ZLZLZF00440) and the Health Bureau Science and Technology
Foundation of Tianjin (grant nos. 2012KZ063 and 2014KZ102).
References
|
1
|
Pawlik TM, Paulino AF, McGinn CJ, Baker
LH, Cohen DS, Morris JS, Rees R and Sondak VK: Cutaneous
angiosarcoma of the scalp: A multidisciplinary approach. Cancer.
98:1716–1726. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Young RJ, Brown NJ, Reed MW, Hughes D and
Woll PJ: Angiosarcoma. Lancet Oncol. 11:983–991. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kuntzen D, Hanel M Tufail, Kuntzen T,
Yurtsever H, Tuma J, Hopfer H, Springer O and Bock A: Malignant
hemangiosarcoma in a renal allograft: Diagnostic difficulties and
clinical course after nephrectomy and immunostimulation. Transpl
Int. 27:e70–e75. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Scow JS, Reynolds CA, Degnim AC, Petersen
IA, Jakub JW and Boughey JC: Primary and secondary angiosarcoma of
the breast: The Mayo Clinic experience. J Surg Oncol. 101:401–407.
2010.PubMed/NCBI
|
|
5
|
Babarović E, Zamolo G, Mustać E and Strčić
M: High grade angiosarcoma arising in fibroadenoma. Diagn Pathol.
6:1252011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Terada T: Angiosarcoma of the mandibular
gingiva. Int J Clin Exp Pathol. 4:791–793. 2011.PubMed/NCBI
|
|
7
|
Sher SJ, Mujtaba MA, Yaqub MS, Taber TE,
Mishler DP and Sharfuddin AA: Early fatal cutaneous lower extremity
angiosarcoma associated with deep venous thrombosis in a renal
transplant recipient. Exp Clin Transplant. Jul 2–2015.(Epub ahead
of print). PubMed/NCBI
|
|
8
|
Kan T, Yanase T, Kawai M and Hide M:
Angiosarcoma of elderly patients treated with weekly low-dose
docetaxel in combination with radiotherapy. Skin Cancer. 28:20–23.
2013. View Article : Google Scholar
|
|
9
|
Stiles JM, Amaya C, Rains S, Diaz D, Pham
R, Battiste J, Modiano JF, Kokta V, Boucheron LE, Mitchell DC and
Bryan BA: Targeting of beta adrenergic receptors results in
therapeutic efficacy against models of hemangioendothelioma and
angiosarcoma. PLoS One. 8:e600212013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Albiñana V, Gómez de Las Heras K Villar,
Serrano-Heras G, Segura T, Perona-Moratalla AB, Mota-Pérez M, de
Campos JM and Botella LM: Propranolol reduces viability and induces
apoptosis in hemangioblastoma cells from von Hippel-Lindau
patients. Orphanet J Rare Dis. 10:1182015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
El Hachem M, Bonamonte D, Diociaiuti A,
Mantuano M and Teruzzi C: Cost-utility analysis of propranolol
versus corticosteroids in the treatment of proliferating infantile
hemangioma in Italy. PharmacoEconomics Italian Res Artic. 17:22015.
View Article : Google Scholar
|
|
12
|
Diboun I, Wernisch L, Orengo CA and
Koltzenburg M: Microarray analysis after RNA amplification can
detect pronounced differences in gene expression using limma. BMC
Genomics. 7:2522006. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Sherman BT, da W Huang, Tan Q, Guo Y, Bour
S, Liu D, Stephens R, Baseler MW, Lane HC and Lempicki RA: DAVID
Knowledgebase: A gene-centered database integrating heterogeneous
gene annotation resources to facilitate high-throughput gene
functional analysis. BMC Bioinformatics. 8:4262007. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Szklarczyk D, Franceschini A, Wyder S,
Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos
A, Tsafou KP, et al: STRING v10: Protein-protein interaction
networks, integrated over the tree of life. Nucleic Acids Res.
43:D447–D452. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Bandettini WP, Kellman P, Mancini C,
Booker OJ, Vasu S, Leung SW, Wilson JR, Shanbhag SM, Chen MY and
Arai AE: MultiContrast delayed enhancement (MCODE) improves
detection of subendocardial myocardial infarction by late
gadolinium enhancement cardiovascular magnetic resonance: A
clinical validation study. J Cardiovasc Magn Reson. 14:832012.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yip KY, Yu H, Kim PM, Schultz M and
Gerstein M: The tYNA platform for comparative interactomics: A web
tool for managing, comparing and mining multiple networks.
Bioinformatics. 22:2968–2970. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Gong H, Xu DP, Li YX, Cheng C, Li G and
Wang XK: Evaluation of the efficacy and safety of propranolol,
timolol maleate, and the combination of the two, in the treatment
of superficial infantile haemangiomas. Br J Oral Maxillofac Surg.
53:836–840. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Oksiuta M, Matuszczak E, Debek W,
Dzienis-Koronkiewicz E and Hermanowicz A: Treatment of rapidly
proliferating haemangiomas in newborns with propranolol and review
of the literature. J Matern Fetal Neonatal Med. 29:64–68. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Praticò AD, Caraci F, Pavone P, Falsaperla
R, Drago F and Ruggieri M: Propranolol: Effectiveness and failure
in infantile cutaneous hemangiomas. Drug Saf Case Rep. 2:62015.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Sanwald R and Wagener H: Enzymes of the
carbohydrate, lipid and protein metabolism in the blood vessels of
the miniature pig. Z Gesamte Exp Med. 145:353–355. 1968.(In
German). View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Stroev IuI, Volgin EG, Bodrov VE and
Chuchman AD: Changes in elastic and viscous properties of blood
vessels in patients with diabetes mellitus and their relationship
with disorders of glycoprotein and lipid metabolism. Kardiologiia.
16:130–132. 1976.(In Russian). PubMed/NCBI
|
|
23
|
Hassan HH, Denis M, Krimbou L, Marcil M
and Genest J: Cellular cholesterol homeostasis in vascular
endothelial cells. Can J Cardiol. 22 Suppl B:B35–B40. 2006.
View Article : Google Scholar
|
|
24
|
Lee CH, Poburko D, Kuo KH, Seow CY and van
Breemen C: Ca(2+) oscillations, gradients, and homeostasis in
vascular smooth muscle. Am J Physiol Heart Circ Physiol.
282:H1571–H1583. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ausprunk DH and Folkman J: Migration and
proliferation of endothelial cells in preformed and newly formed
blood vessels during tumor angiogenesis. Microvasc Res. 14:53–65.
1977. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Anderson M, Ober A, Mollard A, Call L,
Bearss JJ, Vankayalapati H, Sharma S, Warner S and Bearss DJ:
Abstract 5543: Inhibition of the tyrosine kinase receptor Axl
blocks cell invasion and promotes apoptosis in pancreatic cancer
cells. Cancer Res. 73:5543. 2013. View Article : Google Scholar
|
|
27
|
Mudduluru G and Allgayer H: The human
receptor tyrosine kinase Axl gene-promoter characterization and
regulation of constitutive expression by Sp1, Sp3 and CpG
methylation. Biosci Rep. 28:161–176. 2008.PubMed/NCBI
|
|
28
|
Mudduluru G, Vajkoczy P and Allgayer H:
Myeloid zinc finger 1 induces migration, invasion, and in vivo
metastasis through Axl gene expression in solid cancer. Mol Cancer
Res. 8:159–169. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Hafizi S and Dahlbäck B: Signalling and
functional diversity within the Axl subfamily of receptor tyrosine
kinases. Cytokine Growth Factor Rev. 17:295–304. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Axelrod H and Pienta KJ: Axl as a mediator
of cellular growth and survival. Oncotarget. 5:8818–8852. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Rochlitz C, Lohri A, Bacchi M, Schmidt M,
Nagel S, Fopp M, Fey MF, Herrmann R and Neubauer A: Axl expression
is associated with adverse prognosis and with expression of Bcl-2
and CD34 in de novo acute myeloid leukemia (AML): Results from a
multicenter trial of the Swiss group for clinical cancer research
(SAKK). Leukemia. 13:1352–1358. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Challier C, Uphoff CC, Janssen JW and
Drexler HG: Differential expression of the ufo/axl oncogene in
human leukemia-lymphoma cell lines. Leukemia. 10:781–787.
1996.PubMed/NCBI
|
|
33
|
Neubauer A, O'Bryan JP, Fiebeler A,
Schmidt C, Huhn D and Liu ET: Axl, a novel receptor tyrosine kinase
isolated from chronic myelogenous leukemia. Semin Hematol. 30(3
Suppl 3): S341993.
|
|
34
|
Tworkoski K, Singhal G, Szpakowski S, Zito
CI, Bacchiocchi A, Muthusamy V, Bosenberg M, Krauthammer M, Halaban
R and Stern DF: Phosphoproteomic screen identifies potential
therapeutic targets in melanoma. Mol Cancer Res. 9:801–812. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Green J, Ikram M, Vyas J, Patel N, Proby
CM, Ghali L, Leigh IM, O'Toole EA and Storey A: Overexpression of
the Axl tyrosine kinase receptor in cutaneous SCC-derived cell
lines and tumours. Br J Cancer. 94:1446–1451. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Liu L, Greger J, Shi H, Liu Y, Greshock J,
Annan R, Halsey W, Sathe GM, Martin AM and Gilmer TM: Novel
mechanism of lapatinib resistance in HER2-positive breast tumor
cells: Activation of AXL. Cancer Res. 69:6871–6878. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Hong CC, Lay JD, Huang JS, Cheng AL, Tang
JL, Lin MT, Lai GM and Chuang SE: Receptor tyrosine kinase AXL is
induced by chemotherapy drugs and overexpression of AXL confers
drug resistance in acute myeloid leukemia. Cancer Lett.
268:314–324. 2008. View Article : Google Scholar : PubMed/NCBI
|