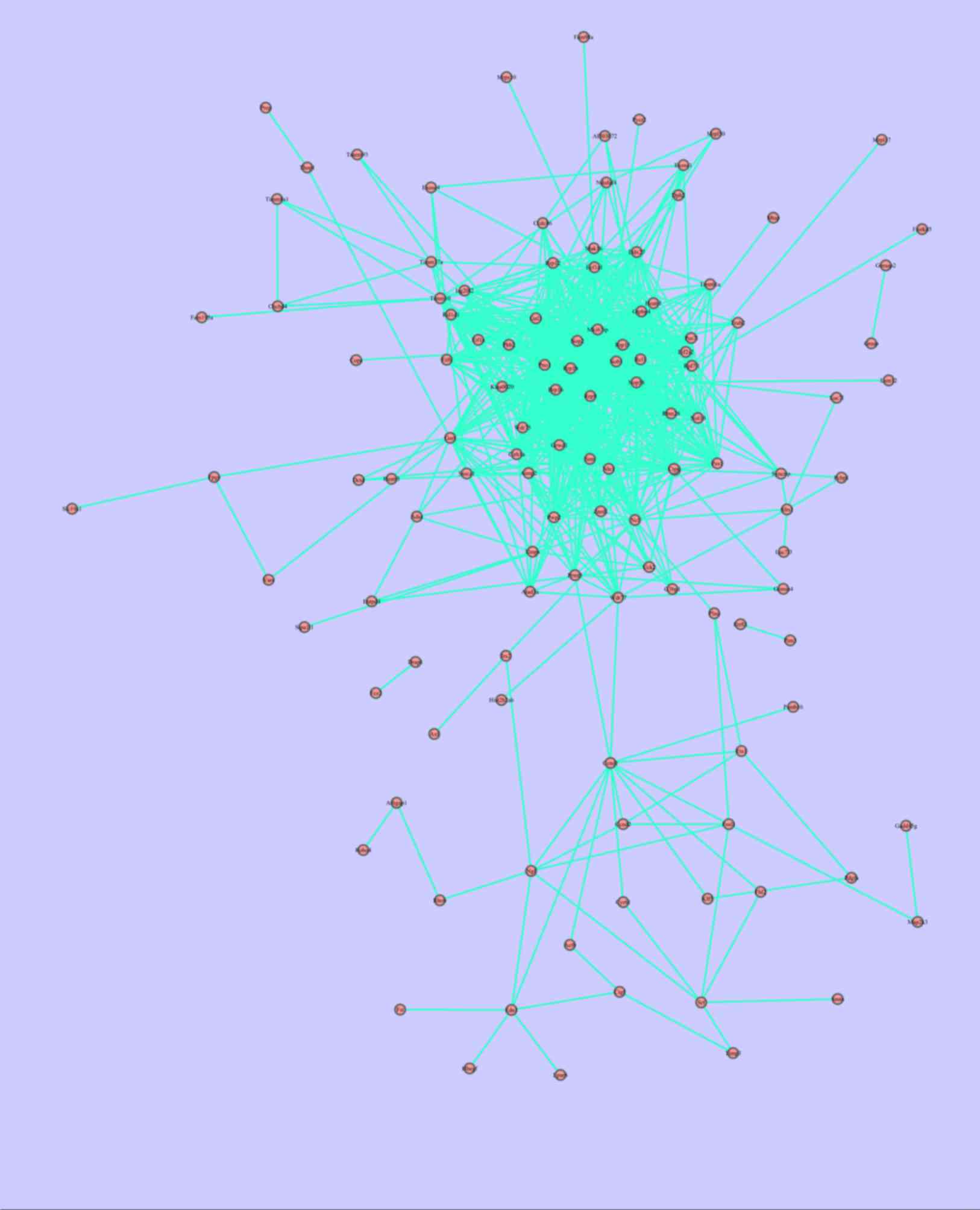
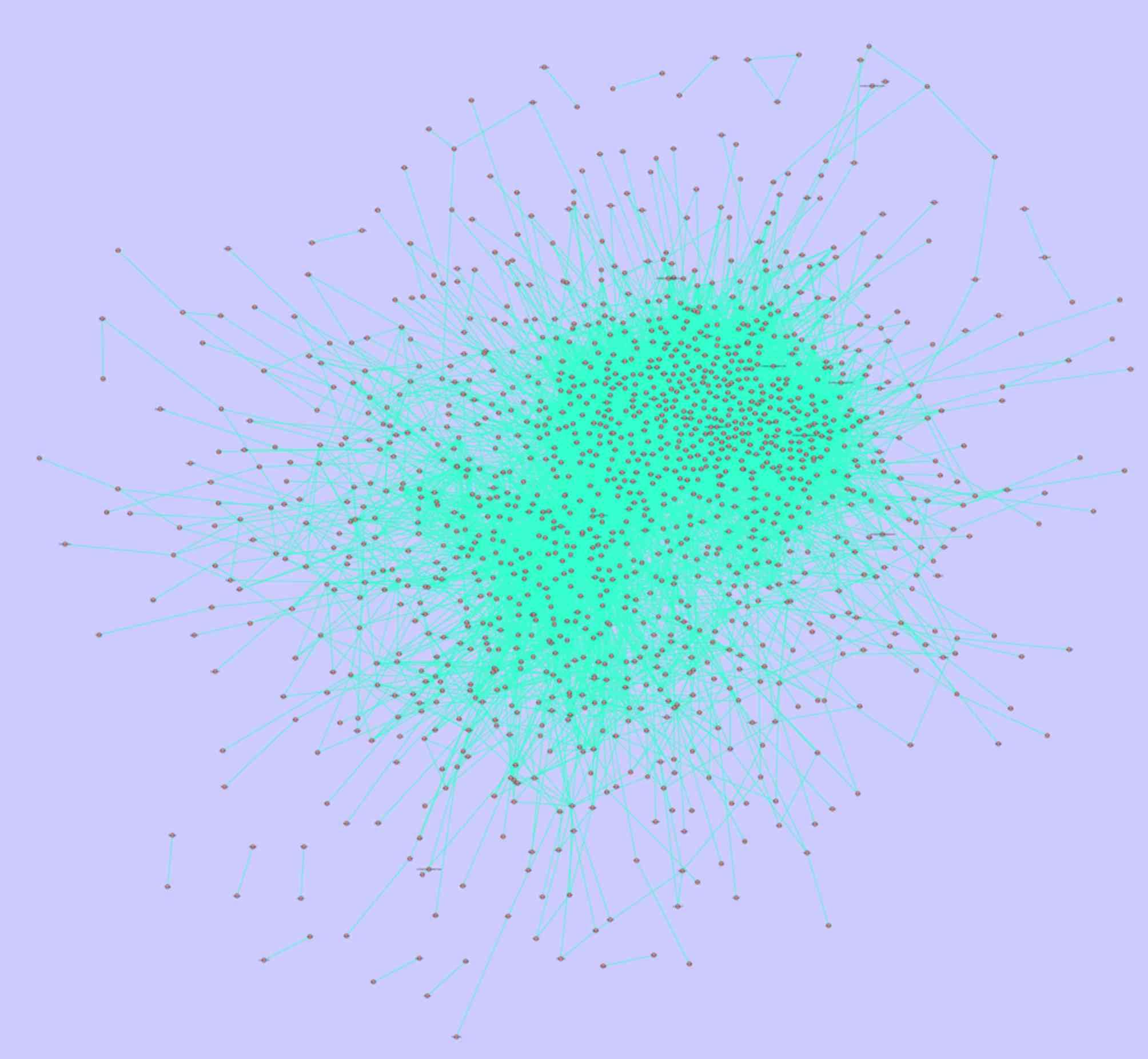
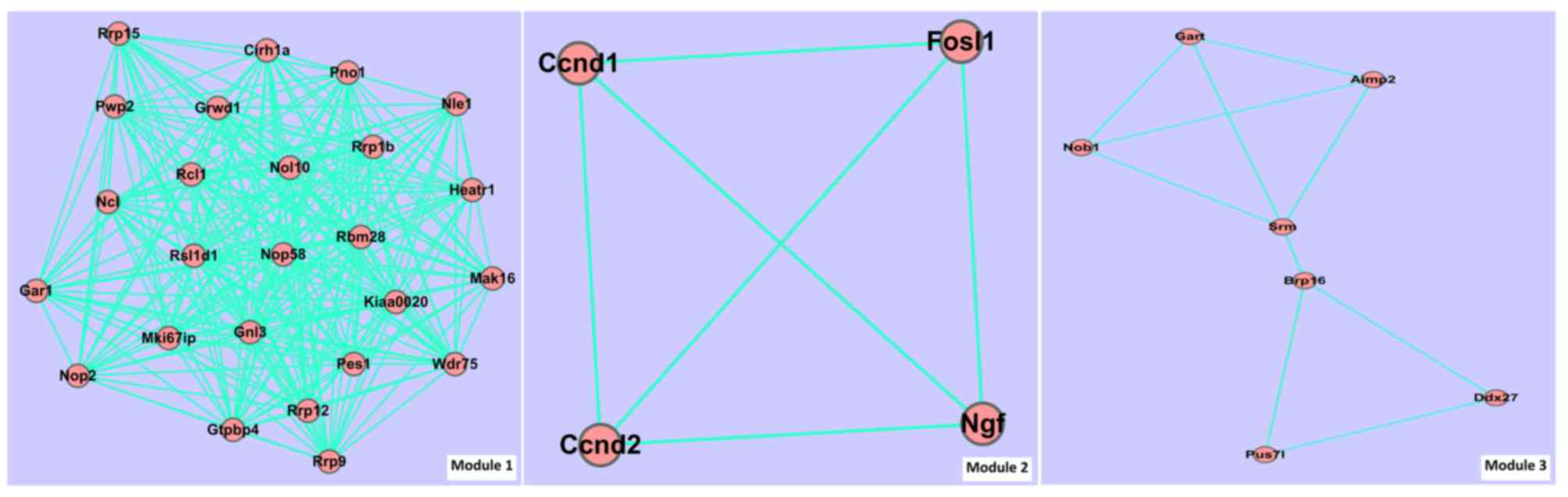
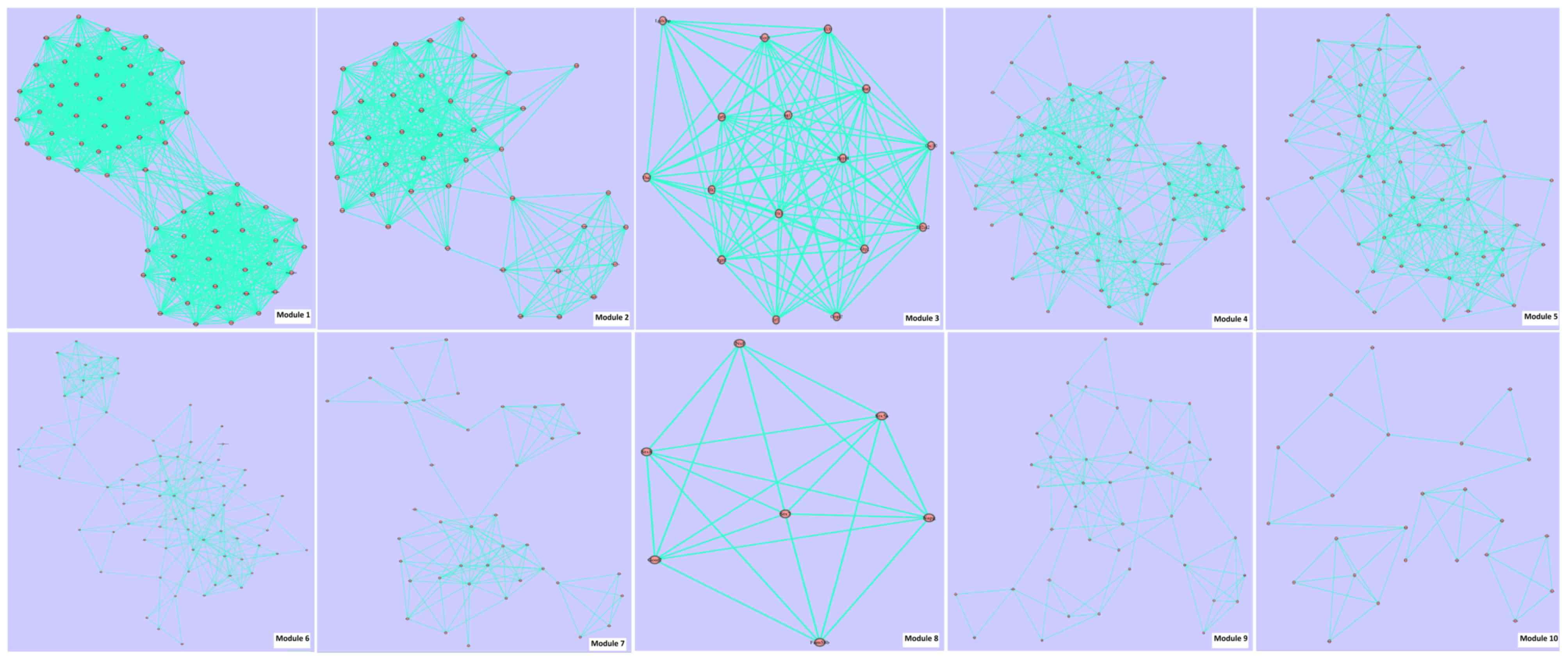
|
1
|
Pawlik TM, Paulino AF, McGinn CJ, Baker
LH, Cohen DS, Morris JS, Rees R and Sondak VK: Cutaneous
angiosarcoma of the scalp: A multidisciplinary approach. Cancer.
98:1716–1726. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Young RJ, Brown NJ, Reed MW, Hughes D and
Woll PJ: Angiosarcoma. Lancet Oncol. 11:983–991. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kuntzen D, Hanel M Tufail, Kuntzen T,
Yurtsever H, Tuma J, Hopfer H, Springer O and Bock A: Malignant
hemangiosarcoma in a renal allograft: Diagnostic difficulties and
clinical course after nephrectomy and immunostimulation. Transpl
Int. 27:e70–e75. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Scow JS, Reynolds CA, Degnim AC, Petersen
IA, Jakub JW and Boughey JC: Primary and secondary angiosarcoma of
the breast: The Mayo Clinic experience. J Surg Oncol. 101:401–407.
2010.PubMed/NCBI
|
|
5
|
Babarović E, Zamolo G, Mustać E and Strčić
M: High grade angiosarcoma arising in fibroadenoma. Diagn Pathol.
6:1252011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Terada T: Angiosarcoma of the mandibular
gingiva. Int J Clin Exp Pathol. 4:791–793. 2011.PubMed/NCBI
|
|
7
|
Sher SJ, Mujtaba MA, Yaqub MS, Taber TE,
Mishler DP and Sharfuddin AA: Early fatal cutaneous lower extremity
angiosarcoma associated with deep venous thrombosis in a renal
transplant recipient. Exp Clin Transplant. Jul 2–2015.(Epub ahead
of print). PubMed/NCBI
|
|
8
|
Kan T, Yanase T, Kawai M and Hide M:
Angiosarcoma of elderly patients treated with weekly low-dose
docetaxel in combination with radiotherapy. Skin Cancer. 28:20–23.
2013. View Article : Google Scholar
|
|
9
|
Stiles JM, Amaya C, Rains S, Diaz D, Pham
R, Battiste J, Modiano JF, Kokta V, Boucheron LE, Mitchell DC and
Bryan BA: Targeting of beta adrenergic receptors results in
therapeutic efficacy against models of hemangioendothelioma and
angiosarcoma. PLoS One. 8:e600212013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Albiñana V, Gómez de Las Heras K Villar,
Serrano-Heras G, Segura T, Perona-Moratalla AB, Mota-Pérez M, de
Campos JM and Botella LM: Propranolol reduces viability and induces
apoptosis in hemangioblastoma cells from von Hippel-Lindau
patients. Orphanet J Rare Dis. 10:1182015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
El Hachem M, Bonamonte D, Diociaiuti A,
Mantuano M and Teruzzi C: Cost-utility analysis of propranolol
versus corticosteroids in the treatment of proliferating infantile
hemangioma in Italy. PharmacoEconomics Italian Res Artic. 17:22015.
View Article : Google Scholar
|
|
12
|
Diboun I, Wernisch L, Orengo CA and
Koltzenburg M: Microarray analysis after RNA amplification can
detect pronounced differences in gene expression using limma. BMC
Genomics. 7:2522006. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Sherman BT, da W Huang, Tan Q, Guo Y, Bour
S, Liu D, Stephens R, Baseler MW, Lane HC and Lempicki RA: DAVID
Knowledgebase: A gene-centered database integrating heterogeneous
gene annotation resources to facilitate high-throughput gene
functional analysis. BMC Bioinformatics. 8:4262007. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Szklarczyk D, Franceschini A, Wyder S,
Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos
A, Tsafou KP, et al: STRING v10: Protein-protein interaction
networks, integrated over the tree of life. Nucleic Acids Res.
43:D447–D452. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Bandettini WP, Kellman P, Mancini C,
Booker OJ, Vasu S, Leung SW, Wilson JR, Shanbhag SM, Chen MY and
Arai AE: MultiContrast delayed enhancement (MCODE) improves
detection of subendocardial myocardial infarction by late
gadolinium enhancement cardiovascular magnetic resonance: A
clinical validation study. J Cardiovasc Magn Reson. 14:832012.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yip KY, Yu H, Kim PM, Schultz M and
Gerstein M: The tYNA platform for comparative interactomics: A web
tool for managing, comparing and mining multiple networks.
Bioinformatics. 22:2968–2970. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Gong H, Xu DP, Li YX, Cheng C, Li G and
Wang XK: Evaluation of the efficacy and safety of propranolol,
timolol maleate, and the combination of the two, in the treatment
of superficial infantile haemangiomas. Br J Oral Maxillofac Surg.
53:836–840. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Oksiuta M, Matuszczak E, Debek W,
Dzienis-Koronkiewicz E and Hermanowicz A: Treatment of rapidly
proliferating haemangiomas in newborns with propranolol and review
of the literature. J Matern Fetal Neonatal Med. 29:64–68. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Praticò AD, Caraci F, Pavone P, Falsaperla
R, Drago F and Ruggieri M: Propranolol: Effectiveness and failure
in infantile cutaneous hemangiomas. Drug Saf Case Rep. 2:62015.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Sanwald R and Wagener H: Enzymes of the
carbohydrate, lipid and protein metabolism in the blood vessels of
the miniature pig. Z Gesamte Exp Med. 145:353–355. 1968.(In
German). View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Stroev IuI, Volgin EG, Bodrov VE and
Chuchman AD: Changes in elastic and viscous properties of blood
vessels in patients with diabetes mellitus and their relationship
with disorders of glycoprotein and lipid metabolism. Kardiologiia.
16:130–132. 1976.(In Russian). PubMed/NCBI
|
|
23
|
Hassan HH, Denis M, Krimbou L, Marcil M
and Genest J: Cellular cholesterol homeostasis in vascular
endothelial cells. Can J Cardiol. 22 Suppl B:B35–B40. 2006.
View Article : Google Scholar
|
|
24
|
Lee CH, Poburko D, Kuo KH, Seow CY and van
Breemen C: Ca(2+) oscillations, gradients, and homeostasis in
vascular smooth muscle. Am J Physiol Heart Circ Physiol.
282:H1571–H1583. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ausprunk DH and Folkman J: Migration and
proliferation of endothelial cells in preformed and newly formed
blood vessels during tumor angiogenesis. Microvasc Res. 14:53–65.
1977. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Anderson M, Ober A, Mollard A, Call L,
Bearss JJ, Vankayalapati H, Sharma S, Warner S and Bearss DJ:
Abstract 5543: Inhibition of the tyrosine kinase receptor Axl
blocks cell invasion and promotes apoptosis in pancreatic cancer
cells. Cancer Res. 73:5543. 2013. View Article : Google Scholar
|
|
27
|
Mudduluru G and Allgayer H: The human
receptor tyrosine kinase Axl gene-promoter characterization and
regulation of constitutive expression by Sp1, Sp3 and CpG
methylation. Biosci Rep. 28:161–176. 2008.PubMed/NCBI
|
|
28
|
Mudduluru G, Vajkoczy P and Allgayer H:
Myeloid zinc finger 1 induces migration, invasion, and in vivo
metastasis through Axl gene expression in solid cancer. Mol Cancer
Res. 8:159–169. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Hafizi S and Dahlbäck B: Signalling and
functional diversity within the Axl subfamily of receptor tyrosine
kinases. Cytokine Growth Factor Rev. 17:295–304. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Axelrod H and Pienta KJ: Axl as a mediator
of cellular growth and survival. Oncotarget. 5:8818–8852. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Rochlitz C, Lohri A, Bacchi M, Schmidt M,
Nagel S, Fopp M, Fey MF, Herrmann R and Neubauer A: Axl expression
is associated with adverse prognosis and with expression of Bcl-2
and CD34 in de novo acute myeloid leukemia (AML): Results from a
multicenter trial of the Swiss group for clinical cancer research
(SAKK). Leukemia. 13:1352–1358. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Challier C, Uphoff CC, Janssen JW and
Drexler HG: Differential expression of the ufo/axl oncogene in
human leukemia-lymphoma cell lines. Leukemia. 10:781–787.
1996.PubMed/NCBI
|
|
33
|
Neubauer A, O'Bryan JP, Fiebeler A,
Schmidt C, Huhn D and Liu ET: Axl, a novel receptor tyrosine kinase
isolated from chronic myelogenous leukemia. Semin Hematol. 30(3
Suppl 3): S341993.
|
|
34
|
Tworkoski K, Singhal G, Szpakowski S, Zito
CI, Bacchiocchi A, Muthusamy V, Bosenberg M, Krauthammer M, Halaban
R and Stern DF: Phosphoproteomic screen identifies potential
therapeutic targets in melanoma. Mol Cancer Res. 9:801–812. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Green J, Ikram M, Vyas J, Patel N, Proby
CM, Ghali L, Leigh IM, O'Toole EA and Storey A: Overexpression of
the Axl tyrosine kinase receptor in cutaneous SCC-derived cell
lines and tumours. Br J Cancer. 94:1446–1451. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Liu L, Greger J, Shi H, Liu Y, Greshock J,
Annan R, Halsey W, Sathe GM, Martin AM and Gilmer TM: Novel
mechanism of lapatinib resistance in HER2-positive breast tumor
cells: Activation of AXL. Cancer Res. 69:6871–6878. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Hong CC, Lay JD, Huang JS, Cheng AL, Tang
JL, Lin MT, Lai GM and Chuang SE: Receptor tyrosine kinase AXL is
induced by chemotherapy drugs and overexpression of AXL confers
drug resistance in acute myeloid leukemia. Cancer Lett.
268:314–324. 2008. View Article : Google Scholar : PubMed/NCBI
|