Introduction
Colorectal carcinoma (CRC) is the third most common
type of cancer worldwide and has increasing rates of incidence in
the Asia-Pacific region, including China (1–3). A number
of studies have shown that a variety of genetic and epigenetic
alterations in both oncogenes and tumor suppressors are involved in
the pathogenesis of CRC. The activation of oncogenes such as the
ras gene and the inactivation of tumor suppressors such as
adenomatous polyposis coli and p53 genes have been well documented
in CRC (4–6). In addition, we previously identified
certain genetic changes, including the downregulation of MUS81
structure-specific endonuclease subunit and epidermal growth
factor-like protein 8 precursor, to be associated with this
malignancy (7,8). However, further investigations are still
necessary to clarify the tumorigenic pathway of CRC (9).
WEE1 is a member of the serine/threonine protein
kinase gene family originally defined by Thuriaux et al
(10) in fission yeast
Schizosaccharomyces pombe. In mammals, WEE1 encodes a 94 kDa
protein with 647 amino acid residues and composes a small gene
family with myelin transcription factor 1 (11,12). WEE1
is located predominantly in the nucleus and selectively
phosphorylates the phospho-cdc2 residue of the cell division cycle
2 and inactivates it (13,14). WEE1 is therefore a critical G2
checkpoint regulator that induces interphase and prevents the
initiation of mitosis (15). Previous
studies have shown that WEE1 expression is increased in various
types of human malignancies including melanoma and vulvar squamous
cell, ovarian and hepatocellular carcinoma (HCC) (16–19).
Elevated WEE1 expression is also documented in glioblastoma and
breast cancer cells, and the inhibition of WEE1 in these cells
results in suppressed cellular proliferation and increased
apoptosis (20–22). In addition, a potent and selective
inhibitor of WEE1 protein, AZD1775, has demonstrated the ability to
sensitize T cell acute lymphoblastic leukemia to cytarabine by
promoting apoptosis over DNA repair (23). AZD1775, formerly termed MK-1775, may
also enhance the therapeutic effects of chemotherapy agents,
including 5-fluorouracil, doxorubicin, camptothecin and mitomycin
C, in various p53-deficient colon cancer cells (24). Although these previous studies have
suggested WEE1 as a promising target in the therapy of human
malignancies including CRC (25), the
data regarding the expression pattern of WEE1 in human CRC tissues
remains limited (26).
The present study therefore detected the expression
levels of the WEE1 gene and protein in CRC tissues by
reverse-transcription quantitative polymerase chain reaction
(RT-qPCR) and immunohistochemistry. The correlations between WEE1
expression and clinicopathological features were also studied, as
well as the prognosis of patients with CRC.
Materials and methods
Patients and specimens
Matched cancerous and adjacent normal tissue
specimens were obtained from 43 patients with CRC who underwent
surgery at Guangzhou Red Cross Hospital (Guangdong, China) between
March 2012 and January 2013. These specimens were collected and
frozen in liquid nitrogen immediately following surgery until
RT-qPCR analysis. Additionally, 102 cases of paraffin-embedded CRC
tissue specimens (without corresponding normal tissues) were also
collected from patients who underwent surgery at the same hospital
between March 2008 and December 2009. Clinicopathological data
including age, gender, tumor size, tumor histology, lymph node
status, tumor node metastasis (TNM) stage and metastasis of all
patients in these two cohorts were recorded. Prognostic data
including tumor free survival time and overall survival time were
also collected for the 102 patients. Prior written informed consent
was obtained from all patients involved in the study and the study
protocol was approved by the Ethics Committee of Guangzhou Red
Cross Hospital. Diagnoses of CRC were all confirmed by
histopathological examination.
RNA preparation and RT
Total RNA was extracted from the 43 CRC tissues and
the corresponding normal tissues using TRIzol reagent (Invitrogen;
Thermo Fisher Scientific, Inc., Waltham, MA, USA) according to the
manufacturer's protocol and as described previously (27). Total RNA was reverse transcribed using
the PrimeScript™ RT reagent kit (Takara Bio, Inc., Kusatsu, Shiga,
Japan) in a final volume of 20 µl containing 4.0 µl of the total
RNA sample, 1.0 µl PrimeScript™ RT enzyme mix I, 1.0 µl of Oligo dT
primer (50 µM), 2.0 µl of random 6-mers (100 µM), 2.0 µl of
5xPrimeScript™ buffer and 10.0 µl of RNase free distilled
H2O. The RT reaction was performed by a C1000 Thermal
cycler (Bio-Rad Laboratories, Inc. Hercules, CA, USA) using the
following conditions: 37°C for 15 min, then 85°C for 5 sec.
TaqMan-based qPCR for WEE1 mRNA
TaqMan-based qPCR was performed on the extracted RNA
using a 7300 Real-Time PCR system (Applied Biosystems; Thermo
Fisher Scientific, Inc.). The PCR assay was carried out in a 25 µl
reaction system consisting of 2.0 µl of cDNA sample, 0.5 µl of each
primer (forward and reverse, 10 pmol/µl), 0.5 µl of Taq DNA
polymerase (Takara Bio, Inc.), 0.5 µl of deoxynucleotide
triphosphate mixture (Takara Bio, Inc.), 0.5 µl of probe (5
pmol/µl), 2.5 µl of 10x PCR buffer (Takara Bio, Inc.) and 18.0 µl
of distilled H2O. The PCR amplification consisted of 40
cycles of 93°C for 15 sec, 55°C for 25 sec and 72°C for 25 sec
subsequent to an initial denaturation step (93°C for 2 min). The
primers and probes for PCR assay were designed by Primer Express
2.0 (Applied Biosystems; Thermo Fisher Scientific, Inc.) and
synthesized by Invitrogen (Thermo Fisher Scientific, Inc.). The
TaqMan probe for WEE1 was 5′-FAM-CTGCTGGTGCTGAACCTCTTCC-BHQ1-3′.
Primers for WEE1 were forward, 5′-GCTTGCCCTCACAGTGGTATG-3′ and
reverse, 5′-CCGAGGTAATCTACCCTGTCT-GA-3′. To correct the differences
in quality and quantity of complementary DNA (cDNA) samples,
β-actin gene was measured in the same samples as an internal
control. The TaqMan probe for β-actin was
5′-FAM-CCTCACCCTGAAGTACCCCAT-CGAGC-BHQ1-3′. Primers for β-actin
were forward, 5′-GCATGGGTCAGAAGGATTC-CT-3′ and reverse,
5′-TCGTCCCAGTTGGTGACGAT-3′. The negative control contained water
instead of cDNA. All samples, including the negative control, were
analyzed in triplicate.
Quantification for PCR products and
score of WEE1 mRNA upregulation
One sample of normal colon mucosa was used as the
calibrator to prepare the standard curves for each gene. The target
gene of the calibrator was amplified using a qPCR assay with the
same conditions as the test samples. The qPCR product was verified
by 2% low melting point agarose gel electrophoresis and
subsequently extracted and purified using a QIAquick Gel Extraction
kit (Qiagen GmbH, Hilden, Germany). The purified product was
measured for optical density (OD) 260 and OD 280 and the purity
value was satisfactory when the OD 260/OD 280 value was >1.8.
The concentration (copy/µl) of the purified product was calculated
according to the OD 260 value and the length of the product.
The purified product with a dilution of 1:10 was
used as the highest concentration point for the construction of the
standard curve. The rest of the standard curve points were prepared
by 4 subsequent serial 10x dilutions. All 5 standards were measured
with the tissue samples under the same conditions, and the
generated standard curve was used to quantify the products of PCR.
The relative expression levels of WEE1 mRNA in CRC tissue samples
were normalized to the internal control β-actin and calculated as
the quantity of WEE1 mRNA (copy/µl)/the quantity of β-actin mRNA
(copy/µl). Score of WEE1 mRNA upregulation=the relative expression
of WEE1 mRNA in cancer tissue/the relative expression of WEE1 mRNA
in the corresponding normal tissue. Score of WEE1 mRNA upregulation
was defined as positive when it was >1.5.
Immunohistochemistry analysis
WEE1 protein expression was detected by
immunohistochemistry analysis in 102 cases of CRC tissues, in which
43 specimens of CRC were not included. The protocol of
immunohistochemistry analysis is described briefly as follows.
Tissue sections of 2 µm thickness were cut and baked at 60°C for 1
h, deparaffinized in xylene and rehydrated through graded ethanol
washes (100% ethanol for 3 min, twice; 95% ethanol for 3 min,
twice; distilled water for 3 min). Subsequently, sections were
subjected to microwave heat-induced antigen retrieval in citrate
buffer (0.01 M, pH 6.0; Beijing Zhongshan Golden Bridge
Biotechnology Co., Ltd., Beijing, China) at high power for 3 min
and cooled to room temperature by the gradual addition of water for
at least 20 min.
Subsequent to rinsing with distilled water, 3%
hydrogen peroxide was applied to block the endogenous peroxidases
at room temperature for 10 min. Sections were then rinsed with PBS
5 times (2 min each time). Samples were incubated at 4°C overnight
with the polyclonal rabbit anti-human WEE1 antibody (dilution,
1:75; catalog no., PAB3322; Abnova, Taipei City, Taiwan).
Subsequent to rinsing 5 times with PBS (2 min each time), the
sections were incubated with anti-rabbit horseradish peroxide
immunoglobulin G (dilution, 1:300; catalog no., SPN-9001; Beijing
Zhongshan Golden Bridge Biotechnology Co., Ltd) for 30 min at 37°C.
Slides were then visualized by applying
3,3-diaminobenzidine-tetrahydrochloride for 4 min and then were
counterstained with hematoxylin.
One case of hepatocellular carcinoma, with
significantly elevated WEE1 protein expression (16), was used as the positive control. The
negative control slide was probed with normal goat serum (Beijing
Zhongshan Golden Bridge Biotechnology Co., Ltd.) under the same
experimental conditions. WEE1 staining was examined by counting 200
cells in the region of interest, which focused on tumor cells. No
significant necrosis was identified by two independent pathologists
who were blind to the clinical characteristics of the samples. The
intensity of WEE1 staining was classified using a 4-point scale
according to previous literature (18): 0, no positive cell; 1+, <10%
positive cells; 2+, 10–50% positive cells; 3+, >50% positive
cells. The expression of WEE1 protein was defined as negative if
the score was 0 and was classified as positive if the scores were
1+, 2+ or 3+. Based on immunohistochemistry results, patients with
CRC were divided into the low WEE1 staining score group (0 or 1+)
and the high WEE1 staining score group (2+ or 3+) to compare the
prognosis between these two groups.
Statistical analysis
Student's t-test was applied to compare the WEE1
mRNA expression levels in 43 cases of CRC tissues with the
corresponding normal tissues. The χ2 test (or Fisher's
exact test, for categorical data) and Student's t-test (for
continuous data) were used to analyze the correlations between the
scores of WEE1 mRNA upregulation in 43 cases of CRC tissue samples
and the clinicopathological characteristics of CRC. The
χ2 test or Fisher's exact test was used to analyze the
associations between WEE1 immunostaining scores in 102 cases of CRC
samples and the clinicopathological characteristics of CRC.
Survival curves were constructed using the Kaplan-Meier method and
the differences in tumor-free survival and overall survival were
evaluated by a Log-rank test. Univariable Cox regression analysis
was employed to explore the prognostic implication of
clinicopathological features, including gender, age, maximal tumor
size, tumor differentiation, depth of tumor invasion, lymph node
metastasis, distant metastasis, TNM stage and WEE1 immunostaining
score. Those clinicopathological features associated with prognosis
of CRC were subsequently put into a multivariable Cox regression
model to identify factors that were independently associated with
overall survival rate of CRC. In this model, a step-wise selection
was used for variable selection with entry and removal limits of
P≤0.05 and P>0.10, respectively. All statistical analyses were
two-sided and performed by SPSS 13.0 software package (SPSS, Inc.,
Chicago, IL, USA). The continuous data were expressed as the mean ±
standard deviation. P<0.05 was considered to indicate a
statistically significant difference.
Results
The upregulation of WEE1 mRNA in CRC
tissues
WEE1 mRNA was detectable in all 43 cases of CRC
tissue specimens and its relative expression level in CRC tissues
was significantly increased compared with the corresponding
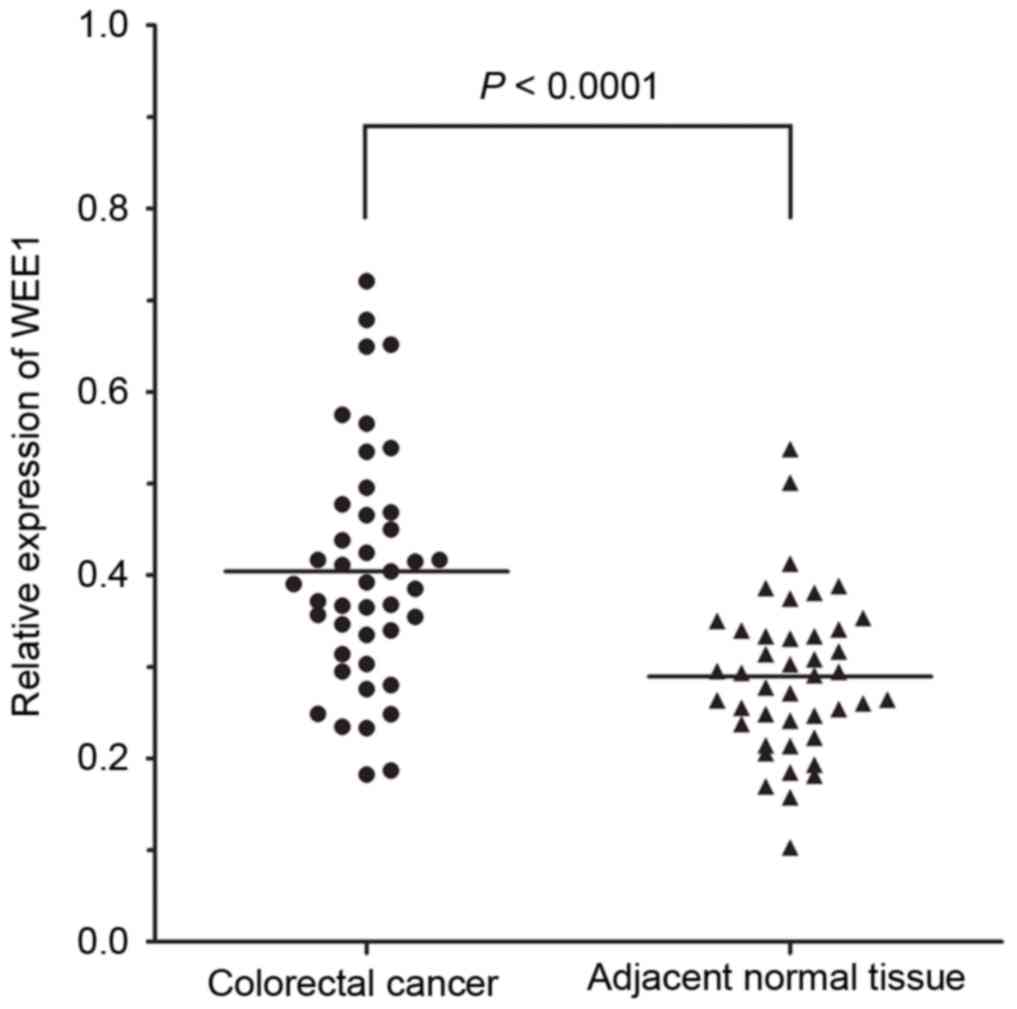
adjacent normal tissues (0.404±0.129 vs. 0.289±0.086, P<0.0001;
Fig. 1). In addition, the score of
WEE1 mRNA upregulation in 43 cases of CRC tissue specimens was
1.519±0.773 and positive in 41.9% (18/43) of specimens.
Association between the scores of WEE1
mRNA upregulation and clinicopathological variables of CRC
To explore the clinical association of WEE1 mRNA
upregulation in CRC, the clinicopathological data was associated
with the scores of WEE1 mRNA upregulation. The present results
demonstrated that the scores of WEE1 mRNA upregulation were
significantly associated to hepatic metastasis (P=0.035), distant
metastasis (P=0.039) and high TNM stage (P=0.039) of patients with
CRC (Table I). However, no
significant association was identified between the scores of WEE1
mRNA upregulation and the other clinicopathological features,
including gender, age, maximal tumor size, tumor differentiation
and lymph node metastasis.
 | Table I.Association between the scores of WEE1
mRNA upregulation and clinicopathological variables of colorectal
cancer. |
Table I.
Association between the scores of WEE1
mRNA upregulation and clinicopathological variables of colorectal
cancer.
|
|
| Upregulation of WEE1
mRNA |
|
|---|
|
|
|
|
|
|---|
| Clinicopathological
variable | n | Positive | Negative | P-value |
|---|
| Gender |
|
|
| 0.234a |
|---|
| Male | 26 | 9 | 17 |
|
Female | 17 | 9 | 8 |
| Age, years | 43 |
64.8±14.6b | 71.1±12.2b | 0.129c |
| Maximal tumor size,
mm | 43 | 52.71±23.8b |
56.9±27.1b | 0.610c |
| Tumor
differentiation |
|
|
| 0.234a |
|
Well-Moderate | 26 | 9 | 17 |
| Poor | 17 | 9 | 8 |
| Lymph node
metastasis |
|
|
| 0.455a |
|
Presence | 21 | 10 | 11 |
|
Absence | 22 | 8 | 14 |
| Hepatic
metastasis |
|
|
| 0.035a |
|
Presence | 8 | 6 | 2 |
|
Absence | 35 | 12 | 23 |
| Distant
metastasis |
|
|
| 0.039a |
|
Presence | 10 | 7 | 3 |
|
Absence | 33 | 11 | 22 |
| Tumor node
metastasis stage |
|
|
| 0.039a |
| I, II,
III | 33 | 11 | 22 |
| IV | 10 | 7 | 3 |
Associations between WEE1
immunostaining scores and clinicopathological variables of CRC
As previously described in melanoma and vulvar
squamous cell carcinoma (17,18), immunohistochemical staining of WEE1
was predominantly detected in the cellular nucleus despite evidence
of positive staining being identified in the cytoplasm. Positive
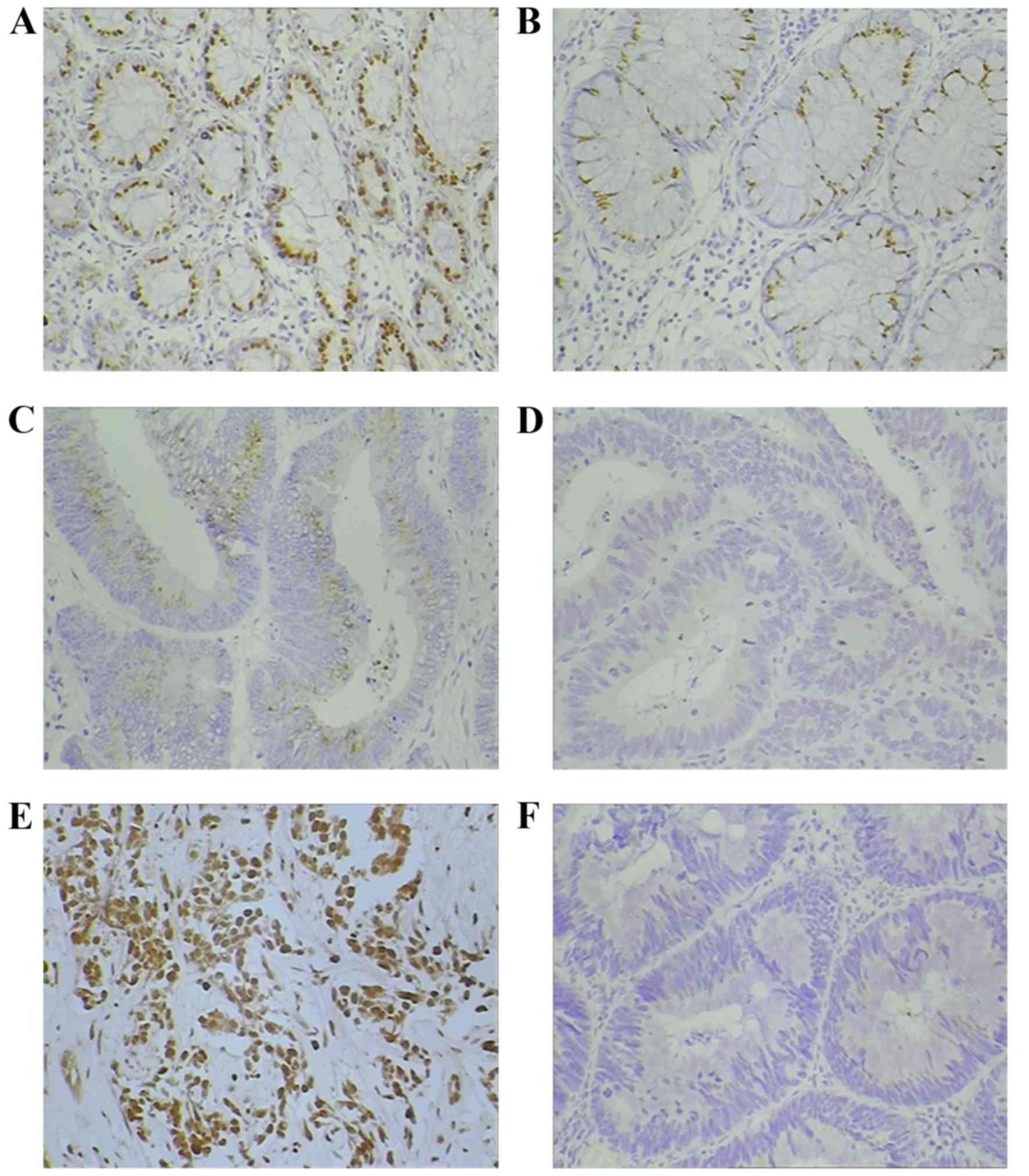
WEE1 expression (Fig. 2A-C) was
evidenced in 52.9% (54/102) of patients with CRC. In patients with
positive WEE1 staining: 10 patients scored 3+; 19 patients scored
2+; and 25 patients scored 1+. However, WEE1 was undetectable in
the remaining 48 patients with CRC and scored 0 (Fig. 2D). When associated with
clinicopathological data, WEE1 staining scores were significantly
correlated to distant metastasis (P=0.002) and a high TNM stage
(P=0.002) of patients with CRC (Table
II). However, no significant association was identified between
WEE1 staining scores and the other clinicopathological features,
including gender, age, maximal tumor size, tumor differentiation,
depth of tumor invasion, and lymph node metastasis, of patients
with CRC.
 | Table II.Association between the WEE1
immunostaining scores and clinicopathological variables of
colorectal cancer. |
Table II.
Association between the WEE1
immunostaining scores and clinicopathological variables of
colorectal cancer.
|
|
| WEE1 immunostaining
scores |
|
|---|
|
|
|
|
|
|---|
| Clinicopathological
variable | n | 0 | 1+ | 2+ | 3+ | P-value |
|---|
| Gender |
|
|
|
|
| 0.607 |
|
Male | 62 | 31 | 16 | 9 | 6 |
|
|
Female | 40 | 17 | 9 | 10 | 4 |
|
| Age, years |
|
|
|
|
| 0.590 |
|
≤65 | 70 | 30 | 18 | 15 | 7 |
|
|
>65 | 32 | 18 | 7 | 4 | 3 |
|
| Maximal tumor
sizea |
|
|
|
|
| 0.351 |
| Mean
≤43.6 mm | 57 | 23 | 17 | 12 | 5 |
|
| Mean
>43.6 mm | 45 | 25 | 8 | 7 | 5 |
|
| Tumor
differentiation |
|
|
|
|
| 0.134 |
|
Well-Moderate | 89 | 43 | 24 | 14 | 8 |
|
|
Poor | 13 | 5 | 1 | 5 | 2 |
|
| Depth of tumor
invasion |
|
|
|
|
| 0.816 |
|
≤Mt | 20 | 9 | 6 | 4 | 1 |
|
|
>Mt | 82 | 39 | 19 | 15 | 9 |
|
| Lymph node
metastasis |
|
|
|
|
| 0.071 |
|
Presence | 30 | 11 | 5 | 9 | 5 |
|
|
Absence | 72 | 37 | 20 | 10 | 5 |
|
| Distant
metastasis |
|
|
|
|
| 0.002 |
|
Presence | 13 | 1 | 3 | 5 | 4 |
|
|
Absence | 89 | 47 | 22 | 14 | 6 |
|
| TNM stage |
|
|
|
|
| 0.002 |
| Stage
I, II, III | 89 | 47 | 22 | 14 | 6 |
|
| Stage
IV | 13 | 1 | 3 | 5 | 4 |
|
Correlations between WEE1 protein
expression and prognosis of patients with CRC
A total of 102 cases of patients with CRC were
divided into two groups based on WEE1 expression levels, the high
WEE1 staining score group (n=29) and the low WEE1 staining score
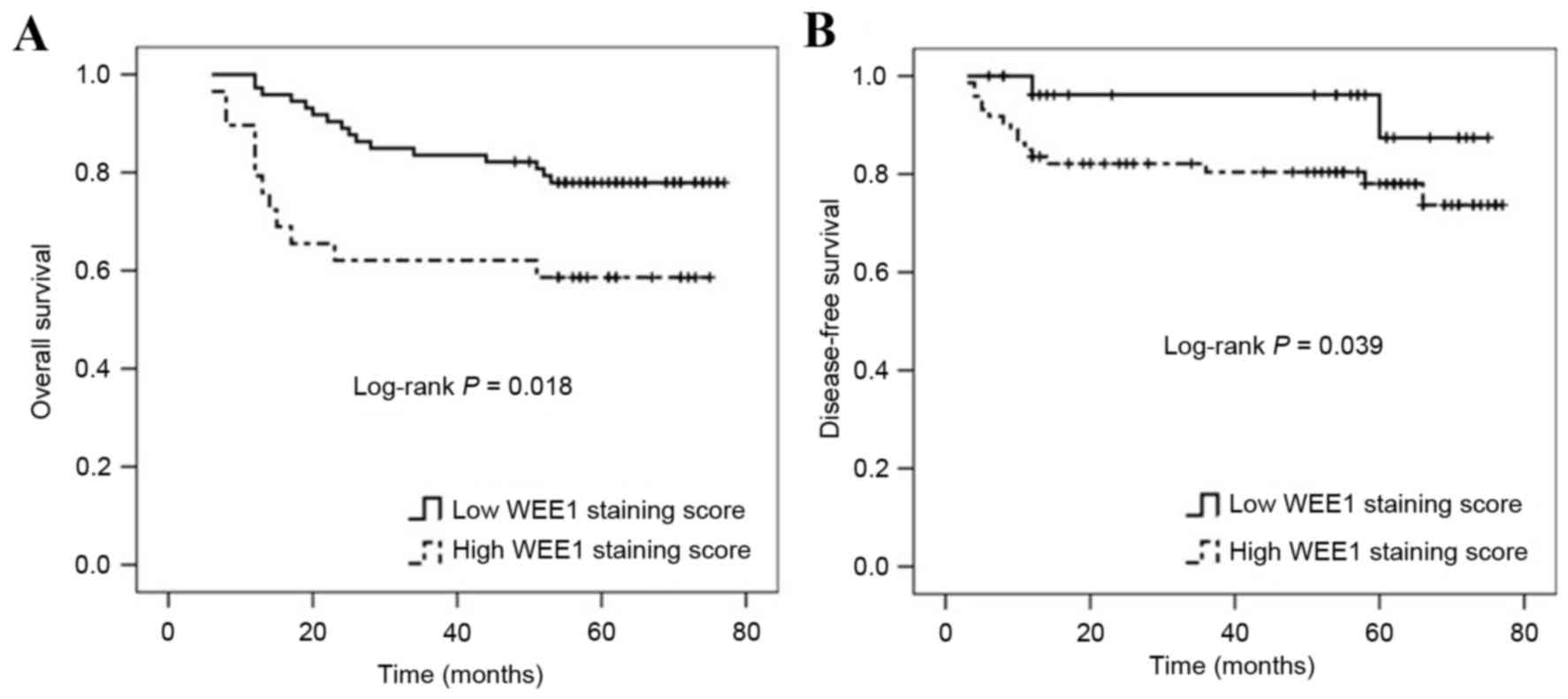
group (n=73). Patients with CRC within the high WEE1 staining score
group had either poorer overall survival (mean overall survival
time, 50.552±5.573 vs. 66.280±2.483 months, P=0.018; Fig. 3A) or poorer tumor-free survival
compared with those within the low WEE1 staining score group (mean
tumor-free survival time, 42.999±3.164 vs. 51.266±2.639 months,
P=0.039; Fig. 3B).
Independent prognostic implication of
high WEE1 staining score for CRC
By using univariable Cox regression analysis,
distant metastasis [hazard ratio (HR), 2.823; P=0.015], high TNM
stage (HR, 4.382; P=0.005), and high WEE1 staining score (HR,
2.392; P=0.023) were associated with the overall survival rate of
patients with CRC (Table III). A
total of 5 variables including gender, age, maximal tumor size,
tumor differentiation, and lymph node metastasis did not enter the
multivariable Cox regression model. It is noteworthy that in this
multivariable model, only high WEE1 expression (HR, 3.339; P=0.039)
and high TNM stage (HR, 5.126; P=0.024) were identified to be
independent prognostic factors for patients with CRC (Table III).
 | Table III.Univariable and multivariable Cox
analysis for the prognostic factors of colorectal cancer. |
Table III.
Univariable and multivariable Cox
analysis for the prognostic factors of colorectal cancer.
|
|
| Univariable
analysis | Multivariable
analysis |
|---|
|
|
|
|
|
|---|
| Variable | n | HR (95% CI) | P-value | HR (95% CI) | P-value |
|---|
| Gender |
|
Male | 62 | 1 | 0.549 | – | – |
|
Female | 40 | 0.797
(0.379–1.379) |
|
|
|
| Age (year,
continuous data) | 102 | 1.117
(0.492–2.492) | 0.791 | – | – |
| Maximal tumor size
(mm, continuous data) | 102 | 0.998
(0.979–1.979) | 0.857 | – | – |
| Tumor
differentiation |
|
Well-mod | 89 | 1 | 0.350 | – | – |
|
Poor | 13 | 1.987
(0.471–8.471) |
|
|
|
| Depth of tumor
invasion |
|
≤Mt | 20 | 1 | 0.596 | – | – |
|
>Mt | 82 | 1.006
(0.512–1.512) |
| LN metastasis |
|
Absence | 72 | 1 | 0.217 | – | – |
|
Presence | 30 | 1.019
(0.989–1.989) |
|
|
|
| Distant
metastasis |
|
Absence | 89 | 1 | 0.015a | 1 | 0.095 |
|
Presence | 13 | 2.823
(1.055–7.055) |
| 3.327
(0.855–9.855) |
|
| TNM stage |
| Stage
I, II, III | 89 | 1 | 0.005a | 1 | 0.024 |
| Stage
IV | 13 | 4.382
(1.595–7.595) |
| 5.126
(1.176–8.176) |
|
| WEE1 staining
score |
| Low
staining score | 73 | 1 | 0.023a | 1 | 0.039 |
| High
staining score | 29 | 2.392
(1.130–5.130) |
| 3.339
(1.030–9.030) |
|
Discussion
Although the upregulation of WEE1 has been observed
in several human malignancies, including in HCC and melanoma, and
in numerous types of tumor cell lines including glioblastoma and
breast cancer (16–19), the data regarding the expression
pattern of WEE1 in human CRC remains limited. Using a cDNA array
and semi-quantitative RT-PCR, Backert et al (26) identified that WEE1 expression was
decreased in human colon cancer cell lines. This downregulation was
verified in 7 cases of CRC tissues despite the evidence that WEE1
was only detectable in 6 cases of normal tissues and 3 cases of CRC
tissue. Considering the difference between cancer tissues and cell
lines, the small number of tissue samples tested and the low
sensitivity and accuracy of conventional RT-PCR, it appears that
the data provided by Backert et al (26) were not enough to ascertain the
expression pattern of WEE1 in CRC tissues. The present study
therefore employed a quantitative TaqMan-based RT-qPCR analysis to
detect the expression levels of WEE1 in 43 cases of CRC tissues and
matched adjacent normal tissues. The present results demonstrated
that WEE1 mRNA was detectable in all CRC tissue and adjacent normal
tissue specimens tested, showing the higher sensitivity of RT-qPCR
compared with conventional RT-PCR. Notably, WEE1 mRNA expression
was significantly increased in CRC tissues compared with the
corresponding adjacent normal tissues and the upregulation of WEE1
mRNA was observed in 41.9% of patients with CRC (18/43 cases). The
upregulation of WEE1 mRNA in CRC is not only in agreement with the
expression pattern of WEE1 in other human malignancy tissues,
including HCC (16–19), but also suggests WEE1 upregulation as
a common event during the carcinogenesis of human malignancies,
including CRC.
Although the exact role of WEE1 in human malignancy
still needs further studies in order to be understood, Magnussen
et al (18) previously
identified that high WEE1 expression is associated with lymph node
metastasis and poor differentiation of vulvar squamous cell
carcinoma. The present study therefore correlated WEE1 mRNA
upregulation with clinicopathological characteristics of CRC and
revealed that the upregulation of WEE1 mRNA was significantly
correlated with the distant metastasis of CRC. In addition, there
was also an association between WEE1 mRNA upregulation and hepatic
metastasis of CRC. Previously, WEE1 expression was increased in
metastatic melanomas compared with primary melanomas (17). Huisman et al (28) also reported that expression rhythm of
WEE1, a circadian clock-controlled gene (29), was completely disrupted in colorectal
liver metastases. Based on these data, the present study presumed
that WEE1 upregulation may be involved in the metastasis potential
of colon cancer, however this should be verified by future studies.
Since distant metastasis is an important parameter in the TNM stage
system of CRC (30), the present
study identified that WEE1 was more strongly upregulated in
patients with CRC with a high TNM stage (stage VI) than those with
a low TNM stage (stage I–III). Together, these results indicate
that upregulation of WEE1 mRNA is closely associated with a high
degree of malignancy in CRC.
Subsequently, the present study determined the
expression pattern of WEE1 protein in 102 cases of CRC by
immunohistochemistry analysis. The present results demonstrated
that WEE1 staining was predominantly observed in the cellular
nucleus although a limited amount of staining was also identified
in cytoplasm, indicating WEE1 as a mainly nucleus-located protein
similar to the location of WEE1 in vulvar squamous cell carcinoma
(18). The present study also
revealed that WEE1 was positive in 52.9% of patients with CRC,
which is lower than the positive rate of WEE1 in melanoma and
vulvar squamous cell carcinoma tissues (17,18) and
suggested the different expression level of WEE1 in different human
tissues. When correlated to clinicopathological data WEE1 protein
staining scores were significantly associated with the distant
metastasis of CRC and high TNM stage. Additionally, there was a
trend in association with WEE1 protein staining scores and lymph
node metastasis of CRC (P=0.071). The present results therefore
increase the evidence for the involvement of WEE1 in the malignancy
progression of CRC.
It has previously been suggested that high WEE1
expression was associated with poor disease-free survival of
malignant melanoma (17). Therefore,
the present study divided the 102 cases of CRC into a low WEE1
expression group and a high WEE1 expression group based on
immunohistochemistry results to explore the prognostic implication
of WEE1 expression. The present results revealed that patients with
CRC within the high WEE1 expression group had either a poorer
disease-free survival or overall survival compared with those
within the low WEE1 expression group. These results are consistent
with results obtained from studies on malignant melanoma,
increasing evidence for the potential prognostic value of high WEE1
expression to CRC. Slipicevic et al (19) identified WEE1 as a novel independent
prognostic marker of poor survival for ovarian carcinoma patients
following chemotherapy by multivariate Cox analysis. Therefore, the
present study also established a Cox regression model, and this
model indicated high WEE1 expression to be an independent risk
factor for the prognosis of patients with CRC.
In conclusion, the present study revealed that WEE1
is upregulated in human CRC tissues and the increased WEE1
expression is correlated with a high degree of malignancy and poor
survival of patients with CRC, which suggests WEE1 as a novel
prognostic marker for CRC. However, further studies are still
required to elucidate the mechanisms underlying the upregulation of
WEE1 in patients with CRC.
Acknowledgements
The authors would like to thank Li-Bing Dai for her
technical assistance in sample collection and storage. The present
study was supported by the Natural Science Foundation of China
(grant no. 81000989), the Natural Science Foundation of Guangdong
Province (grant nos. 10451022002004562 and 2014A030313654), the
Project of Science & Technology New Star of Pearl River by
Guangzhou City (grant no. 2011J2200008), the Fundamental Research
Funds for the Central Universities (grant no. 21615484) and the
Special Support Projection for High-Level Talents of Guangdong
Province (grant no. 2014TQ01R482).
References
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Sung JJ, Lau JY, Young GP, Sano Y, Chiu
HM, Byeon JS, Yeoh KG, Goh KL, Sollano J, Rerknimitr R, et al: Asia
Pacific consensus recommendations for colorectal cancer screening.
Gut. 57:1166–1176. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Matsuda T, Marugame T, Kamo K, Katanoda K,
Ajiki W and Sobue T; Japan Cancer Surveillance Research Group, :
Cancer incidence and incidence rates in Japan in 2006: Based on
data from 15 population-based cancer registries in the monitoring
of cancer incidence in Japan (MCIJ) project. Jpn J Clin Oncol.
42:139–147. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bos JL, Fearon ER, Hamilton SR, Verlaan-de
Vries M, Van Boom JH, van der Eb AJ and Vogelstein B: Prevalence of
ras gene mutations in human colorectal cancers. Nature.
327:293–297. 1987. View
Article : Google Scholar : PubMed/NCBI
|
|
5
|
Baker SJ, Markowitz S, Fearon ER, Willson
JK and Vogelstein B: Suppression of human colorectal carcinoma cell
growth by wild-type p53. Science. 249:912–925. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Nishisho I, Nakamura Y, Miyoshi Y, Miki Y,
Ando H, Horii A, Koyama K, Utsunomiya J, Baba S and Hedge P:
Mutations of chromosome 5q21 genes in FAP and colorectal cancer
patients. Science. 253:665–669. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wu F, Shirahata A, Sakuraba K, Kitamura Y,
Goto T, Saito M, Ishibashi K, Kigawa G, Nemoto H, Sanada Y and Hibi
K: Downregulation of Mus81 as a novel prognostic biomarker for
patients with colorectal carcinoma. Cancer Sci. 102:472–477. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wu F, Shirahata A, Sakuraba K, Kitamura Y,
Goto T, Saito M, Ishibashi K, Kigawa G, Nemoto H, Sanada Y and Hibi
K: Down-regulation of EGFL8: A novel biomarker for advanced gastric
cancer. Anticancer Res. 31:3377–3380. 2011.PubMed/NCBI
|
|
9
|
Walther A, Johnstone E, Swanton C, Midgley
R, Tomlinson I and Kerr D: Genetic prognostic and predictive
markers in colorectal cancer. Nat Rev Cancer. 9:489–499. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Thuriaux P, Nurse P and Carter B: Mutants
altered in the control co-ordinating cell division with cell growth
in the fission yeast Schizosaccharomyces pombe. Mol Gen Genet.
161:215–220. 1978.PubMed/NCBI
|
|
11
|
Perry JA and Kornbluth S: Cdc25 and Wee1:
Analogous opposites? Cell Div. 2:122007. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Mueller PR, Coleman TR, Kumagai A and
Dunphy WG: Myt1: A membrane-associated inhibitory kinase that
phosphorylates Cdc2 on both threonine-14 and tyrosine-15. Science.
270:86–90. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Parker LL and Piwnica-Worms H:
Inactivation of the p34cdc2-cyclin B complex by the human WEE1
tyrosine kinase. Science. 257:1955–1957. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Watanabe N, Broome M and Hunter T:
Regulation of the human WEE1Hu CDK tyrosine 15-kinase during the
cell cycle. EBMO J. 14:1878–1891. 1995.
|
|
15
|
Kawabe T: G2 checkpoint abrogators as
anticancer drugs. Mol Cancer Ther. 3:513–519. 2004.PubMed/NCBI
|
|
16
|
Masaki T, Shiratori Y, Rengifo W, Igarashi
K, Yamagata M, Kurokohchi K, Uchida N, Miyauchi Y, Yoshiji H,
Watanabe S, et al: Cyclins and cyclin-dependent kinases:
Comparative study of hepatocellular carcinoma versus cirrhosis.
Hepatology. 37:534–543. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Magnussen GI, Holm R, Emilsen E, Rosnes
AK, Slipicevic A and Flørenes VA: High expression of Wee1 is
associated with poor disease-free survival in malignant melanoma:
Potential for targeted therapy. PLoS One. 7:e382542012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Magnussen GI, Hellesylt E, Nesland JM,
Trope CG, Flørenes VA and Holm R: High expression of wee1 is
associated with malignancy in vulvar squamous cell carcinoma
patients. BMC Cancer. 13:2882013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Slipicevic A, Holth A, Hellesylt E, Tropé
CG, Davidson B and Flørenes VA: Wee1 is a novel independent
prognostic marker of poor survival in post-chemotherapy ovarian
carcinoma effusions. Gynecol Oncol. 135:118–124. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Mir SE, De Witt Hamer PC, Krawczyk PM,
Balaj L, Claes A, Niers JM, Van Tilborg AA, Zwinderman AH, Geerts
D, Kaspers GJ, et al: In silico analysis of kinase expression
identifies WEE1 as a gatekeeper against mitotic catastrophe in
glioblastoma. Cancer Cell. 18:244–257. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Iorns E, Lord CJ, Grigoriadis A, McDonald
S, Fenwick K, Mackay A, Mein CA, Natrajan R, Savage K, Tamber N, et
al: Integrated functional, gene expression and genomic analysis for
the identification of cancer targets. PLoS One. 4:e51202009.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Murrow LM, Garimella SV, Jones TL, Caplen
NJ and Lipkowitz S: Identification of WEE1 as a potential molecular
target in cancer cells by RNAi screening of the human tyrosine
kinome. Breast Cancer Res Treat. 122:347–357. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ford JB, Baturin D, Burleson TM, Van
Linden AA, Kim YM and Porter CC: AZD1775 sensitizes T cell acute
lymphoblastic leukemia cells to cytarabine by promoting apoptosis
over DNA repair. Oncotarget. 6:28001–28010. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Hirai H, Arai T, Okada M, Nishibata T,
Kobayashi M, Sakai N, Imagaki K, Ohtani J, Sakai T, Yoshizumi T, et
al: MK-1775, a small molecule Wee1 inhibitor, enhances anti-tumor
efficacy of various DNA-damaging agents, including 5-fluorouracil.
Cancer Biol Ther. 9:514–522. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Weisberg E, Nonami A, Chen Z, Liu F, Zhang
J, Sattler M, Nelson E, Cowens K, Christie AL, Mitsiades C, Wong
KK, et al: Identification of Wee1 as a novel therapeutic target for
mutant RAS-driven acute leukemia and other malignancies. Leukemia.
29:27–37. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Backert S, Gelos M, Kobalz U, Hanski ML,
Böhm C, Mann B, Lövin N, Gratchev A, Mansmann U, Moyer MP, et al:
Differential gene expression in colon carcinoma cells and tissues
detected with a cDNA array. Int J Cancer. 82:868–874. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wu F, Liu SY, Tao YM, Ou DP, Fang F and
Yang LY: Decreased expression of methyl methansulfonate and
ultraviolet-sensitive gene clone 81 (Mus81) is correlated with a
poor prognosis in patients with hepatocellular carcinoma. Cancer.
112:2002–2010. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Huisman SA, Oklejewicz M, Ahmadi AR,
Tamanini F, Ijzermans JN, van der Horst GT and de Bruin RW:
Colorectal liver metastases with a disrupted circadian rhythm phase
shift the peripheral clock in liver and kidney. Int J Cancer.
136:1024–1032. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Karantanos T, Theodoropoulos G, Pektasides
D and Gazouli M: Clock genes: Their role in colorectal cancer.
World J Gastroenterol. 20:1986–1992. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zlobec I and Lugli A: Prognostic and
predictive factors in colorectal cancer. J Clin Pathol. 61:561–569.
2008.PubMed/NCBI
|