Introduction
Nasopharyngeal carcinoma (NPC) is the most common
type of malignancy of the neck and head in China (1,2). Due to
the anatomical location, the nasopharynx is difficult to access and
the early symptoms of NPC are not distinctive. Therefore, NPC is
usually diagnosed at the advanced stage (3,4). At
present, the main treatment of NPC is radiotherapy. Radiotherapy
may inhibit the development of NPC, but it may seriously influence
the quality of life of the patient due to the side effects
(5–7).
The regulatory mechanisms of NPC remain unclear. Therefore,
research on the molecular mechanisms and genetic alterations
involved in the progression of NPC is required, as this may help to
find novel therapeutic and diagnostic methods (8–11).
Long non-coding (lnc)RNAs are non-protein-coding
transcripts with >200 nucleotides (12). As the non-coding RNAs do not serve as
the template for protein synthesis, they are considered superfluous
in transcription. Multiple previous studies have demonstrated that
lncRNAs take part in the development of numerous diseases,
particularly via gene silencing, cell cycle regulating, chromatin
remodeling and splicing regulating (13–15). The
lncRNAs are categorized into oncogenes and tumor suppressor genes
according to their different functions in variety of types of
malignancy (16,17). It was demonstrated that the
dysfunctionally expressed lncRNAs were associated with the
occurrence of tumors (18).
Therefore, lncRNAs may be used for the early diagnosis of tumors
and as novel targets for treatments (19,20).
lncRNA metastasis associated lung adenocarcinoma transcript 1 has
already been used as a biomarker for the early diagnosis of
non-small cell lung cancer, due to its specificity compared with
the other serum-specific antigens (21).
lncRNA C22orf32-1 (OTTHUMG00000150697) was
identified through gene expression profile analysis (22). The expression of lncRNA C22orf32-1 was
significantly upregulated in primary NPC tissues compared with the
normal nasopharyngeal epithelial tissues, and therefore may act as
an independent biomarker for early diagnosis and provide a novel
therapeutic target for NPC (13,22,23). In
the present study, the expression levels of lncRNA C22orf32-1 in 24
primary NPC tissues and 24 normal nasopharyngeal epithelial tissues
were examined with one NPC and one normal nasopharyngeal epithelial
cell line. The effect of lncRNA C22orf32-1 knockdown on NPC cell
proliferation, migration, invasion and apoptosis was explored.
Materials and methods
Tissue specimens
A total of 48 samples were collected from Peking
University Shenzhen Hospital (Shenzhen, China) from May 2013 to
February 2015. Of these, 24 samples were NPC tissues from patients
with primary NPC and others were normal nasopharyngeal epithelial
from volunteers without cancer. Fresh samples of normal
nasopharyngeal epithelium and NPC tissues were obtained from the
Department of Head and Neck (Peking University Shenzhen Hospital,
Shenzhen, China), and stored immediately in liquid nitrogen. The
present study was approved by the Research Ethics Board at Peking
University Shenzhen Hospital (Shenzhen, China). No NPC patients
received radiotherapy or chemotherapy prior to the surgery. The
clinical staging of all patients were determined using the 2010
Cancer Staging Standard published by the America Joint Committee
(24). Details of the patient
clinical information are summarized in Table I.
 | Table I.Clinicopathological characteristics
of patients. |
Table I.
Clinicopathological characteristics
of patients.
|
Characteristics | Number of
cases |
|---|
| Mean age, range
(years) | 43 (25–64) |
| Sex
(male/female) | 18/6 |
| Degree of
differentiation (undifferentiated/differentiated) | 15/7 |
| Histology
(squamous/others) | 24/0 |
| Lymph node
metastasis (+/−) | 17/7 |
| Distal metastasis
(+/−) | 0/24 |
| Clinical TNM stage
(I–II/III–IV) | 4/20 |
Cell culture
The normal human nasopharyngeal NP460 and NPC 6–10B
cell lines were obtained from the Southern Medical University
(Guangzhou, China) and Peking University Shenzhen Hospital
(Shenzhen, China). Cells were grown in a humidified incubator with
5% CO2 at 37°C and maintained in medium containing 88%
RPMI-1,640 medium (Gibco; Thermo Fisher Scientific, Inc., Waltham,
MA, USA), 10% fetal bovine serum (FBS; Gibco; Thermo Fisher
Scientific, Inc.), 50 U/ml streptomycin and 50 U/ml penicillin
(Gibco; Thermo Fisher Scientific, Inc.).
RNA extraction, complimentary (c)DNA
synthesis and quantitative reverse transcription polymerase chain
reaction (RT-qPCR)
The RNA was extracted from samples and cell lines
using the TRIzol reagent (Invitrogen; Thermo Fisher Scientific,
Inc.) according to the protocol of the manufacturer. The purity of
RNA was examined in the A260/A280 ratio using the SmartSpec Plus
Spectrophotometer (Bio-Rad Laboratories, Inc., Hercules, USA). The
cDNA was synthesized from RNA using a Reverse Transcription kit
(Takara Biotechnology Co., Ltd., Dalian, China). The lncRNA
C22orf32-1 expression levels in samples and cell lines were
detected with the SYBR Green kit (Takara Biotechnology Co., Ltd.),
and analyzed using the Roche Lightcycler 480 Real-Time PCR System
(Roche Applied Science, Penzberg, Germany). The thermocycling
conditions for RT-qPCR were: 94°C for 3 min, then 94°C for 30 sec,
55°C for 30 sec and 72°C for 2 min for 40 cycles and finally 72°C
for 10 min. All samples were tested in triplicate. Glyceraldehyde
3-phosphate dehydrogenase (GAPDH) was set as the internal control
to normalize the relative expression level of lncRNA C22orf32-1.
The primer sequences were: Forward, 5′-AGCACTTGGCCCTAAAGAGA−3′;
reverse, 5′-AACATACTGGCCCAAACAGC−3′ for lncRNA C22orf32-1, and
forward, 5′-GAGCACAGAGCCTCGCCTTT-3′; reverse,
5′-TCATCATCCATGGTGAGCTGGC-3′ for GAPDH. The expression levels of
lncRNA C22orf32-1 and GAPDH in specimens were calculated using the
ΔΔCq method [ΔΔCq=(meanCqlncRNA
C22orf32-1(NPC)-meanCqGAPDH
(NPC))-(meanCqlncRNA
C22orf32-1(normal)-meanCqGAPDH(normal))]. The
relative RNA expression in cells was calculated using the
2−ΔΔCq method (25).
Small interfering (si)RNA and
transfection of cell lines
A total of four siRNAs against lncRNA C22orf32-1,
si-C22orf32-1, and one negative control, si-NC, were designed and
synthesized by GenePharma (GenePharma, Shanghai, China). All the
four si-C22orf32-1 were: Sense, 5′-CCCAGAGUCACUUAGAAGATT-3′ and
antisense, 5′-AUGGCCAUCAAGAUUAGGGTT-3′; sense,
5′-GAGGCGUGGAGUCUUGUUUTT-3′ and antisense,
5′-AAACAAGACUCCACGCCUCTT-3′; sense, 5′-GGCGGAUUCAUUACAGUUATT-3′ and
antisense, 5′-UAACUGUAAUGAAUCCGCCTT-3′; sense,
5′-CUGGAAUACAAUGCUCUAUTT-3′ and antisense,
5′-AUAGAGCAUUGUAUUCCAGTT-3′. A density of 1×105 NPC
cells were seeded onto 6-well plates overnight and transfected with
100 nM si-C22orf32-1 or 100 nM si-NC using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.). The interfering efficiency was tested using
RT-qPCR 48 h following transfection. The most effective siRNA
against si-C22orf32-1, which had a silence efficiency of >57.3%,
was then selected for further analysis. The primers used were:
Si-C22orf32-1 sense: 5′-CCCUAAUCUUGAUGGCCAUTT-3′ and antisense:
5′-AUGGCCAUCAAGAUUAGGGTT-3′; si-NC sense:
5′-GAGGCGUGGAGUCUUGUUUTT-3′ and antisense:
5′-AAACAAGACUCCACGCCUCTT-3′.
Cell proliferation assays
The cell proliferation assay was conducted with Cell
Counting Kit-8 (CCK8; Dojindo Molecular Technologies, Inc.,
Kumamoto, Japan). A total of 3,000 6–10B cells/well were seeded
onto 10 replicate wells of 96-well plates. A total of 5-wells were
transfected with si-NC and the others were transfected with
si-C22orf32-1. Cell proliferation was monitored every 24 h
following transfection up to 72 h, according to the protocol of the
manufacturer. All assays were performed at least three times.
Cell scratch assay
The cell scratch assay was used to detect the
migration of the NPC cells. A total of 2×106 cells/well
of the 6–10B cell line were plated onto 6-well plates. The cells
were transfected with si-NC and si-C22orf32-1 at 90–100%
confluence. Sterile 200 µl pipette tips were used to scrape a clear
line through the well 6 h following transfection. The serum-free
RPMI-1640 medium was used instead of the previous medium. The
migration distance was assessed and images were captured at 0 and
24 h following scraping using an inverted microscope (Olympus
Corporation, Tokyo, Japan). All experiments were performed in
triplicate.
Transwell and matrigel assays
6-10B cells were harvested 24 h following
transfection with si-C22orf32-1 or si-NC. For the migration assays,
1×104 cells diluted in 100 µl serum-free RPMI-1640
medium were plated into the upper chamber with a pore size 8 µm (BD
Biosciences, Franklin Lakes, NJ, USA). For the invasion assays,
1×104 cells diluted in 100 µl serum-free RPMI-1640
medium were plated into the upper chamber, which was coated with
Matrigel (BD Biosciences, San Jose, CA, USA). A total of 500 µl
medium containing 10% FBS was added to the lower chambers.
Following culture for 24 h, the cells that had invaded and migrated
to the membrane were fixed with paraformaldehyde for 25 min, then
stained with 0.1% crystal violet for 25 min. The images were
captured and cells were counted using an inverted microscope
(Olympus Corporation) and 5 fields of view were selected to be
assessed. All experiments were performed in triplicate.
Cell apoptosis assay
6-10B cells were plated onto 6-well plates and
cultured in a humidified chamber supplemented with 5%
CO2 at 37°C following transfection. The cells were
collected 48 h subsequent to this using EDTA-free pancreatic
enzymes (Gibco; Thermo Fisher Scientific, Inc.) and washed three
times with pre-chilled PBS. Subsequent to resuspension in 1X
binding buffer (Invitrogen; Thermo Fisher Scientific, Inc.), the
cells were stained with a propidium iodide detection kit
(Invitrogen; Thermo Fisher Scientific, Inc.) and Annexin
V-fluorescein isothiocyanate (Invitrogen; Thermo Fisher Scientific)
in the dark for 15 min. Flow cytometry (EPICS XI-4; Beckman
Coulter, Inc., Brea, CA, USA) was used to quantify the rate of
apoptosis. All experiments were performed at least three times.
Statistical analysis
All experiments were performed at least three times,
therefore the data were calculated as the mean ± standard
deviation. All statistical analyses were performed using SPSS
software version 17.0 (SPSS, Inc., Chicago, IL, USA). An unpaired
t-test was to analyze the expression difference between NPC
specimens and normal specimens, and the NPC and the normal
nasopharyngeal epithelial cell lines. Student's t test was used to
analyze the data from the proliferation, invasion, migration,
apoptosis and scratch assays. P<0.05 (two-tailed) was considered
to indicate a statistically significant difference.
Results
The expression of lncRNA C22-orf32-1
is up-regulated in NPC cell lines and human NPC clinical
tissues
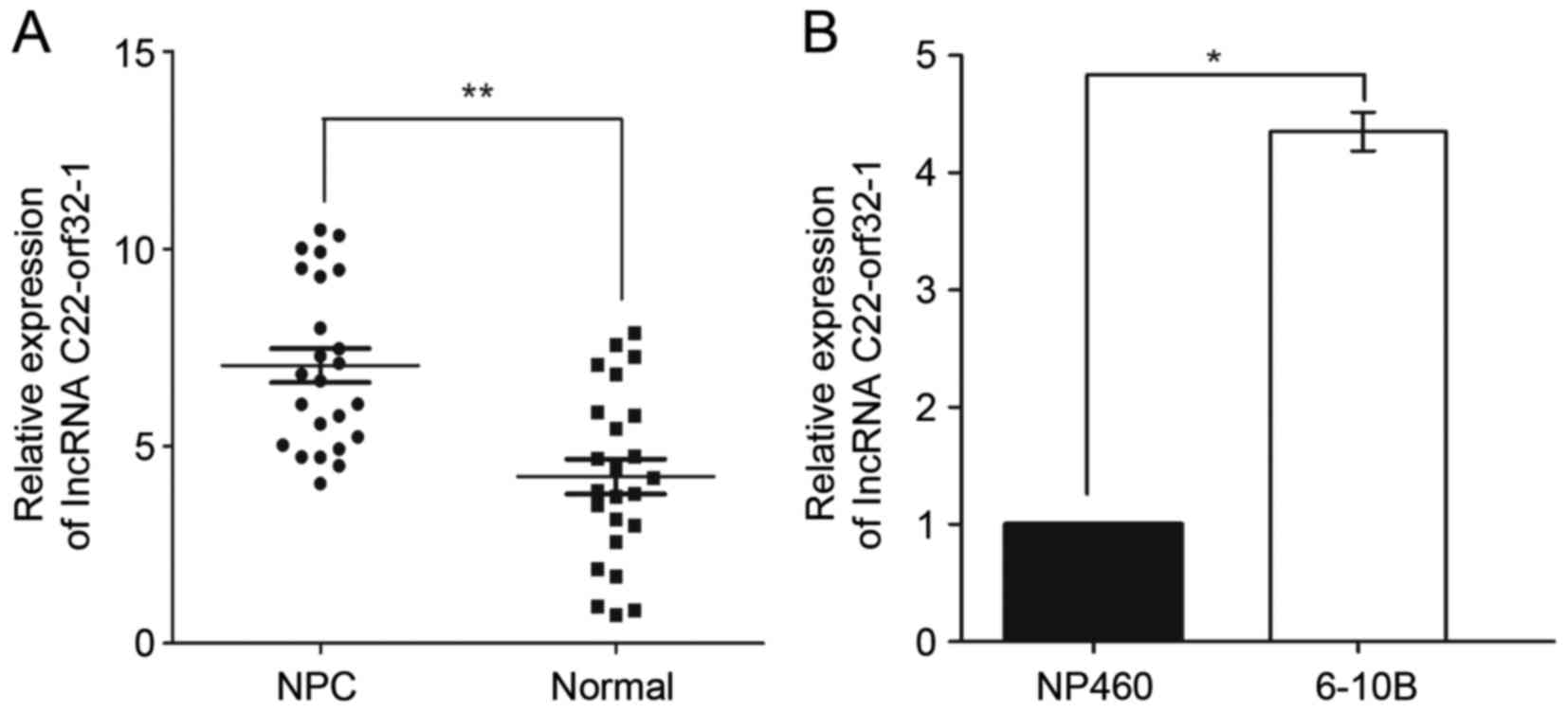
The expression levels of lncRNA C22orf32-1 were
tested in 24 normal nasopharyngeal epithelial tissues and 24 NPC
tissues. The results revealed that lncRNA C22orf32-1 was
significantly upregulated in NPC tissues compared with normal
nasopharyngeal epithelial tissues (Fig.
1A). The NPC 6–10B cell line and normal nasopharyngeal
epithelial NP460 cell line were selected to verify the expression
pattern of lncRNA C22orf32-1. The expression level of lncRNA
C22orf32-1 in the 6–10B cell line was significantly higher compared
with the NP460 cells (Fig. 1B).
Knockdown of lncRNA C22-orf32-1
inhibited the proliferation of NPC cells
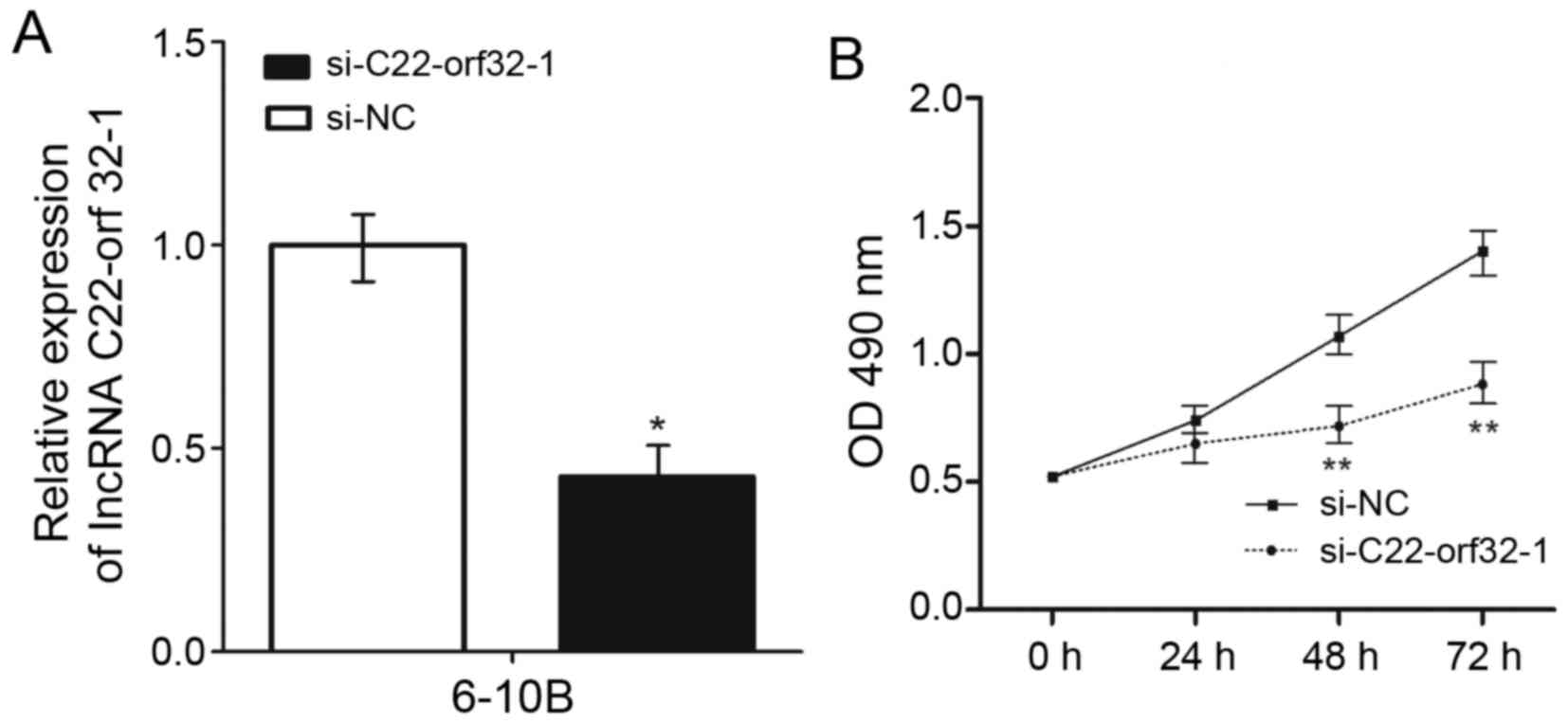
To investigate the potential involvement of lncRNA
C22orf32-1 in NPC progression, four siRNAs were used to decrease
the expression of lncRNA C22orf32-1. The most effective siRNA was
si-C22orf32-1, and further analysis revealed that it significantly
decreased the expression levels of lncRNA C22orf32-1 in the 6–10B
cell lines by >57.3% (P<0.05; Fig.
2A). CCK-8 assays were used to examine the effect of lncRNA
C22orf32-1 on the proliferation of NPC cells. Knockdown of lncRNA
C22orf32-1 significantly suppressed the growth of the 6–10B cell
line 48 and 72 h following transfection compared with the negative
control (Fig. 2B).
LncRNA C22orf32-1 promoted NPC cell
migration and invasion
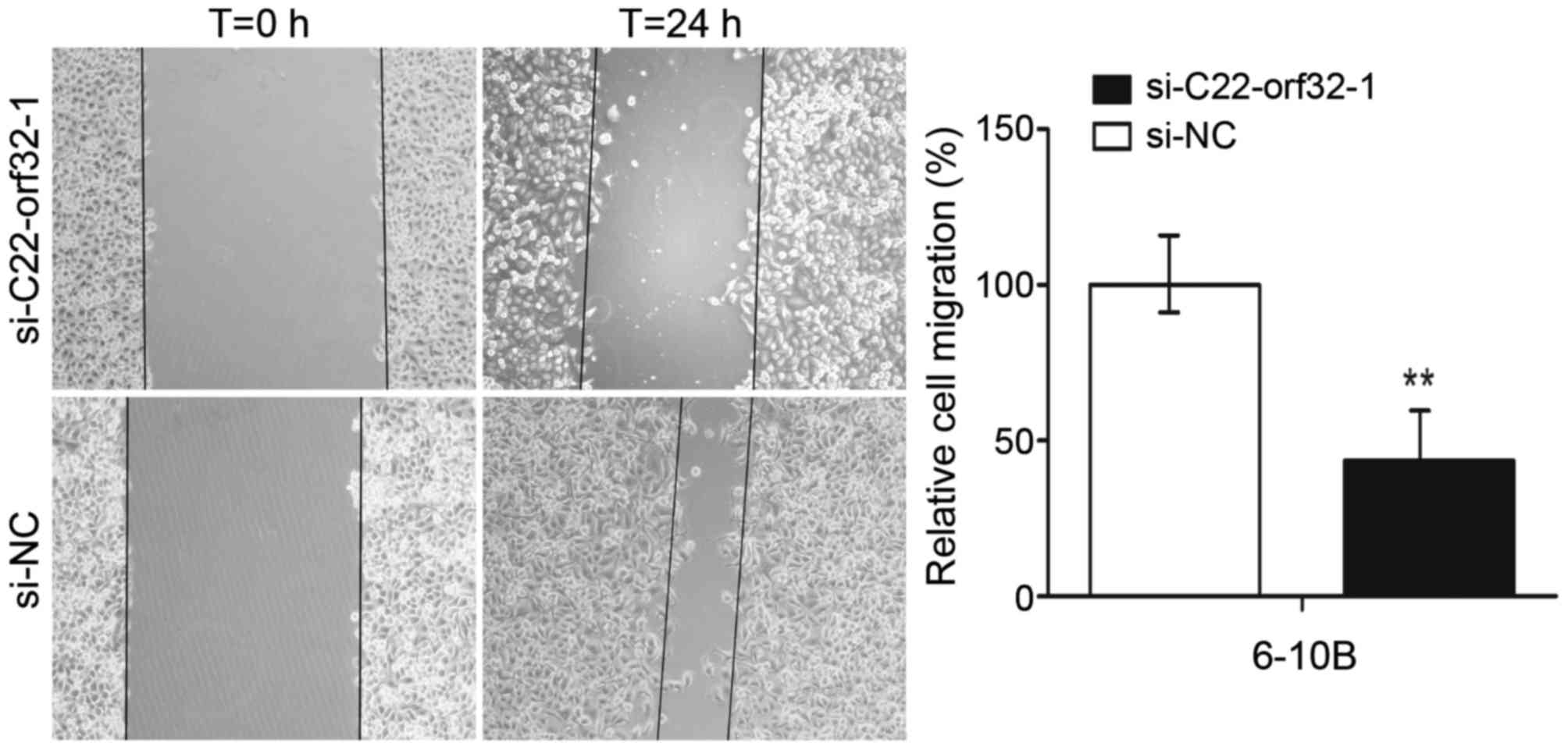
The scratch assays and Transwell migration assays
were used to detect the effect of lncRNA C22orf32-1 on the
migratory ability of NPC cells. In the scratch assays, it was
revealed that the migration abilities of the cells transfected with
si-C22orf32-1 were significantly reduced compared with the cells
transfected with si-NC. The migration rates were reduced to 58% in
6–10B cells (P<0.001; Fig. 3).
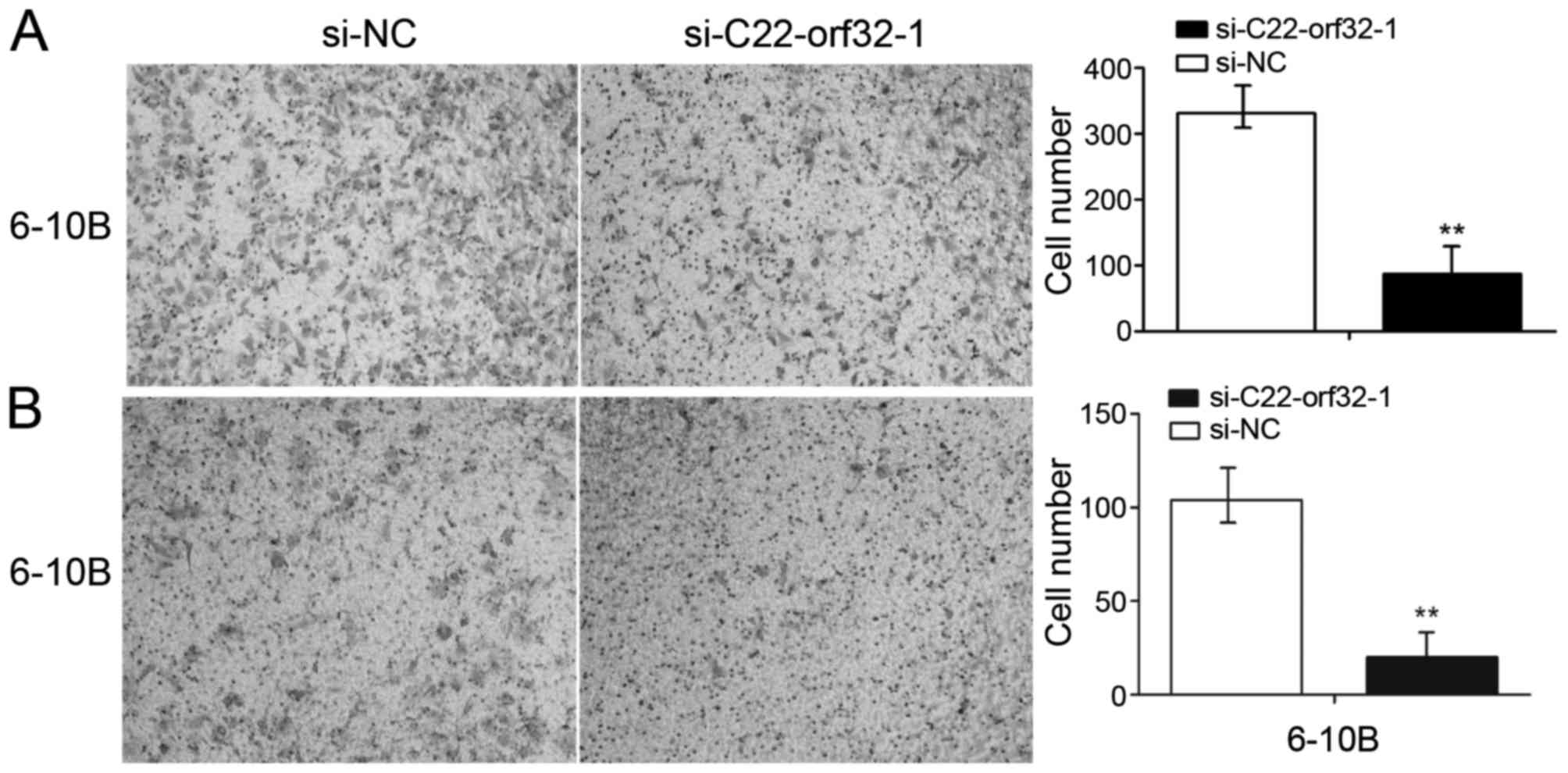
Similarly, the migration rates of the 6–10B cell lines following
transfection with si-C22orf32-1 were decreased by 64.3%
(P<0.001; Fig. 4A) compared with
the negative control. The Matrigel invasion assays were performed
to investigate whether lncRNA C22orf32-1 was involved in the
invasion of NPC cells. The results revealed that lncRNA C22orf32-1
knockdown cells exhibited impeded invasion abilities in the 6–10B
cell line, and were reduced by 76.2% (P<0.001; Fig. 4B).
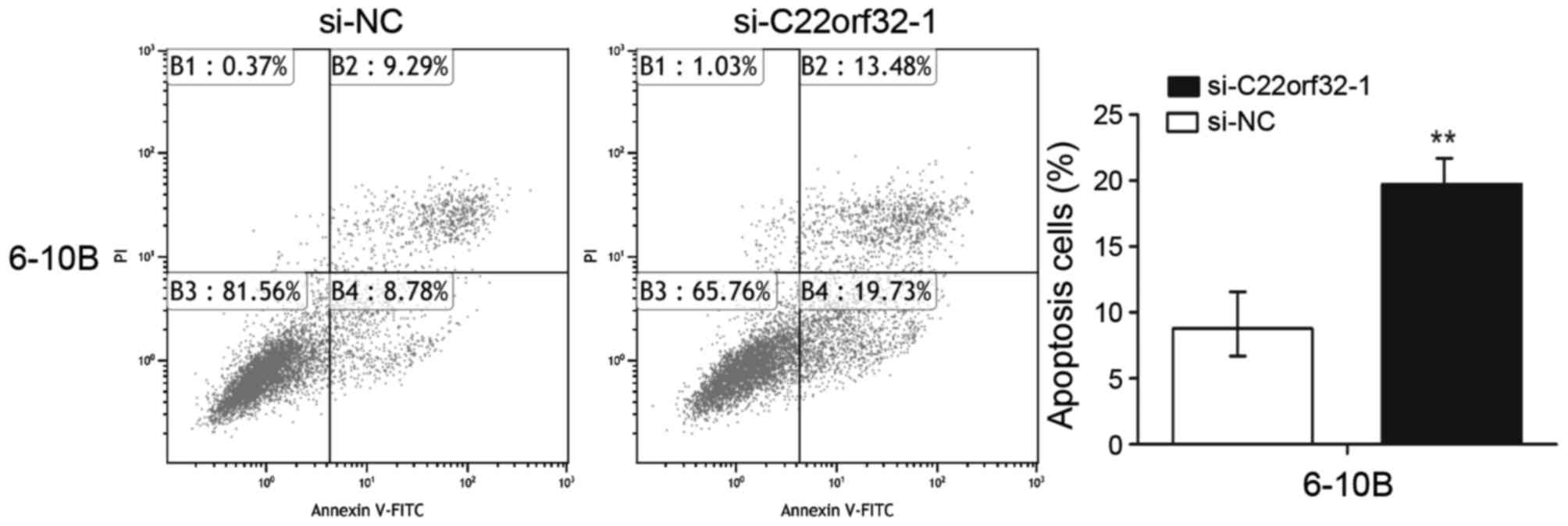
Knockdown of lncRNA C22orf32-1
promoted NPC cell apoptosis
To investigate whether lncRNA C22orf32-1 was
involved in the progression of NPC cell apoptosis, flow cytometry
was used to analysis the rates of apoptosis in the 6–10B cells. The
flow cytometry assay results indicated that the levels of apoptosis
in the cells transfected with si-C22orf32-1 were significantly
increased compared with the cells transfected with si-NC, with 8.78
vs. 19.73% (P<0.001; Fig. 5).
Discussion
Epidemiological studies have demonstrated that NPC
is a specific regional genetic disease with multiple factors.
Exposure to the Epstein-Barr virus, the environment and genetics
are the three main factors that influence the development of NPC
(23,26–28). Like
multiple other human malignancies, the occurrence of NPC is due to
the activation of oncogenes and/or the inactivation of tumor
suppressor genes that disrupt the homeostasis of cellular gene
expression.
LncRNA is considered an important factor that
influences the development of human tumors (29,30).
Previous studies have demonstrated that abnormal expression of
lncRNAs was able to change the biological functions of tumor cells
by affecting various cellular processes. According to their
alternative expression pattern, certain lncRNAs may reflect the
progress of disease, and may be regarded as independent predictors
for cancer diagnosis (14,16,31,32). The
overexpression of Hox transcript antisense RNA in breast cancer
promoted metastasis (33,34) and homeobox A transcript at the distal
tip is considered an indicator for determining the progress of
pancreatic cancer (35). Prostate
cancer associated transcript 1 identified in hepatocellular
carcinoma has been associated with the prognosis in patients with
hepatocellular carcinoma (36). Yet,
the specific functions of lncRNA in the occurrence of tumors
remains unclear.
As demonstrated in a previous study, the expression
of lncRNA C22orf32-1 was upregulated in primary NPC tissues
(22). However, the mechanism of
lncRNA C22orf32-1 in the development of NPC remains unknown
(22). In the present study, the
expression of lncRNA C22orf32-1 was examined in 24 primary NPC
tissues and 24 normal nasopharyngeal epithelial tissues, and in one
NPC and one normal nasopharyngeal epithelial cell line. It was
revealed that the expression levels of lncRNA C22orf32-1 were
higher in the NPC tissues and the NPC cell line. lncRNA C22orf32-1
is located in the human chromosome 22 with a length of 545 bp.
Genetic diversity is a feature of chromosome 22, and as such this
chromosome is associated with the occurrence of human disease. At
present, at least 27 types of disease are associated with
chromosome 22, particularly malignancies including acute lymphoid
leukemia, chronic myelogenous leukemia and malignant rhabdoid
tumors (37–39). The high expression level and the
specific location of lncRNA C22orf32-1 indicate that it may be
associated with NPC. The effect of lncRNA C22orf32-1 knockdown on
NPC cells was investigated, and it was demonstrated that the
overexpression of lncRNA C22orf32-1 in the NPC cell line promoted
cell proliferation, migration, invasion, and suppressed cell
apoptosis. Conversely, subsequent to lncRNA C22orf32-1 knockdown,
the capacities for proliferation, migration and invasion were
decreased, and apoptosis was increased. This indicated that lncRNA
C22orf32-1 is a cancer-associated gene involved in NPC, which may
be a useful candidate biomarker for the early detection and
treatment of NPC. Additional studies are required to determine the
potential mechanisms of lncRNA C22orf32-1 in the development of
NPC.
Acknowledgements
The present study was supported by the Science and
Technology Development Fund Project of Shenzhen (grant nos.
JCYJ20120827150357364, JCYJ20130402114702127 and
JCYJ2015040309144336) and the Medical Research Project of the
Health and Family Planning Commission of Shenzhen (grant no.
201302005).
References
|
1
|
Agulnik M and Epstein JB: Nasopharyngeal
carcinoma: Current management, future directions and dental
implications. Oral Oncol. 44:617–627. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Lee AW, Lin JC and Ng WT: Current
management of nasopharyngeal cancer. Semin Radiat Oncol.
22:233–244. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Spano JP, Busson P, Atlan D, Bourhis J,
Pignon JP, Esteban C and Armand JP: Nasopharyngeal carcinomas: An
update. Eur J Cancer. 39:2121–2135. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Vokes EE, Liebowitz DN and Weichselbaum
RR: Nasopharyngeal carcinoma. Lancet. 350:1087–1091. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
August M, Dodson TB, Nastri A and Chuang
SK: Nasopharyngeal carcinoma: Clinical assessment and review of 176
cases. Oral Surg Oral Med Oral Pathol Oral Radiol Endod.
91:205–214. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Chen W and Hu GH: Biomarkers for enhancing
the radiosensitivity of nasopharyngeal carcinoma. Cancer Biol Med.
12:23–32. 2015.PubMed/NCBI
|
|
7
|
Wolff HA, Rödel RM, Gunawan B, Overbeck T,
Herrmann MK, Hennies S, Hille A, Vorwerk H, Matthias C, Hess CF and
Christiansen H: Nasopharyngeal carcinoma in adults: Treatment
results after long-term follow-up with special reference to
adjuvant interferon-beta in undifferentiated carcinomas. J Cancer
Res Clin Oncol. 136:89–97. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Chou J, Lin YC, Kim J, You L, Xu Z, He B
and Jablons DM: Nasopharyngeal carcinoma-review of the molecular
mechanisms of tumorigenesis. Head Neck. 30:946–963. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Chen ZT, Liang ZG and Zhu XD: A Review:
Proteomics in nasopharyngeal carcinoma. Int J Mol Sci.
16:15497–15530. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Razak AR, Siu LL, Liu FF, Ito E,
O'Sullivan B and Chan K: Nasopharyngeal carcinoma: The next
challenges. Eur J Cancer. 46:1967–1978. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Tulalamba W and Janvilisri T:
Nasopharyngeal carcinoma signaling pathway: An update on molecular
biomarkers. Int J Cell Biol. 2012:5946812012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Morris KV and Mattick JS: The rise of
regulatory RNA. Nat Rev Genet. 15:423–437. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yarmishyn AA and Kurochkin IV: Long
noncoding RNAs: A potential novel class of cancer biomarkers. Front
Genet. 6:1452015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ulitsky I and Bartel DP: lincRNAs:
Genomics, evolution, and mechanisms. Cell. 154:26–46. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Saxena A and Carninci P: Long non-coding
RNA modifies chromatin: Epigenetic silencing by long non-coding
RNAs. Bioessays. 33:830–839. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhang W, Wang L, Zheng F, Zou R, Xie C,
Guo Q, Hu Q, Chen J, Yang X, Yao H, et al: Long noncoding RNA
expression signatures of metastatic nasopharyngeal carcinoma and
their prognostic value. Biomed Res Int. 2015:6189242015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Gibb EA, Brown CJ and Lam WL: The
functional role of long non-coding RNA in human carcinomas. Mol
Cancer. 10:382011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Maruyama R and Suzuli H: Long noncoding
RNA involvement in cancer. BMB Rep. 45:604–611. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Xu T, Su B, Wang C, Wang S, Huang H, Pan
Y, Wang D, Wei W, Claret FX and Yang H: Molecular markers to assess
short-term disease local recurrence in nasopharyngeal carcinoma.
Oncol Rep. 33:1418–1426. 2015.PubMed/NCBI
|
|
20
|
Su YJ, Yu J, Huang YQ and Yang J:
Circulating long noncoding RNA as a potential target for prostate
cancer. Int J Mol Sci. 16:13322–13338. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Weber DG, Johnen G, Casjens S, Bryk O,
Pesch B, Jöckel KH, Kollmeier J and Brüning T: Evaluation of long
noncoding RNA MALAT1 as a candidate blood-based biomarker for the
diagnosis of non-small cell lung cancer. BMC Res Notes. 6:5182013.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Gao W, Chan JY and Wong TS: Differential
expression of long noncoding RNA in primary and recurrent
nasopharyngeal carcinoma. Biomed Res Int. 2014:4045672014.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhou X, Cui J, Macias V, Kajdacsy-Balla
AA, Ye H, Wang J and Rao PN: The progress on genetic analysis of
nasopharyngeal carcinoma. Comp Funct Genomics. 2007:575132007.
View Article : Google Scholar
|
|
24
|
Edge SB and Compton CC: The America joint
committee on cancer: The 7th edition of the AJCC cancer staging
manual and the future of TNM. Ann Surg Oncol. 17:1471–1474. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Chang ET and Adami HO: The enigmatic
epidemiology of nasopharyngeal carcinoma. Cancer Epidemiol
Biomarkers Prev. 15:1765–1777. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kimura Y, Suzuki D, Tokunaga T,
Takabayashi T, Yamada T, Wakisaka N, Yoshizaki T, Murata H, Miwa K,
Shoujaku H, et al: Epidemiological analysis of nasopharyngeal
carcinoma in the central region of Japan during the period from
1996 to 2005. Auris Nasus Larynx. 38:244–249. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Young LS and Dawson CW: Epstein-Barr virus
and nasopharyngeal carcinoma. Chin J Cancer. 33:581–590.
2014.PubMed/NCBI
|
|
29
|
Wapinski O and Chang HY: Long noncoding
RNAs and human disease. Trends Cell Biol. 21:354–361. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Qiu MT, Hu JW, Yin R and Xu L: Long
noncoding RNA: An emerging paradigm of cancer research. Tumour
Biol. 34:613–620. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Quan M, Chen J and Zhang D: Exploring the
secrets of long noncoding RNAs. Int J Mol Sci. 16:5467–5496. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Rinn JL and Chang HY: Genome regulation by
long noncoding RNAs. Annu Rev Biochem. 81:145–166. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Yao Y, Li J and Wang L: Large intervening
non-coding RNA HOTAIR is an indicator of poor prognosis and a
therapeutic target in human cancers. Int J Mol Sci. 15:18985–18999.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zhang J, Zhang P, Wang L, Piao HL and Ma
L: Long non-coding RNA HOTAIR in carcinogenesis and metastasis.
Acta Biochim Biophys Sin (Shanghai). 46:1–5. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Li Z, Zhao X, Zhou Y, Liu Y, Zhou Q, Ye H,
Wang Y, Zeng J, Song Y, Gao W, et al: The long non-coding RNA
HOTTIP promotes progression and gemcitabine resistance by
regulating HOXA13 in pancreatic cancer. J Transl Med. 13:842015.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Yan TH, Yang H, Jiang JH, Lu SW, Peng CX,
Que HX, Lu WL and Mao JF: Prognostic significance of long
non-coding RNA PCAT-1 expression in human hepatocellular carcinoma.
Int J Clin Exp Pathol. 8:4126–4131. 2015.PubMed/NCBI
|
|
37
|
De Decker HP and Lawrenson JB: The 22q11.2
deletion: From diversity to a single gene theory. Genet Med. 3:2–5.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Brinchi V, Curatolo P, Di Franco C and
Dallapiccola B: 2 new cases of chromosome 22 with ring structure.
Pathologica. 71:3691979.(In Italian). PubMed/NCBI
|
|
39
|
Schneider M and Eliez S: 22q11.2
microdeletion. Arch Pediatr. 17:431–434. 2010.(In French).
View Article : Google Scholar : PubMed/NCBI
|