Introduction
Esophageal cancer is the world's eighth most-common
cancer, and the sixth cause of cancer death. Although the morbidity
of Barrett glandular cancer is increasing rapidly in Western
countries, esophageal squamous cell carcinoma (ESCC) still occupies
a leading position in East Asia and China (1). ESCC is often diagnosed at later stage.
Although there are a variety of treatment methods, such as surgery,
chemotherapy and radiotherapy, the prognosis is not satisfactory
(2).
MicroRNA (miRNA or miR) is a recently discovered,
small (approximately 18–24 nucleotides in length) and non-coding
single-stranded RNA that monitors and regulates gene expression
(3). miRNA is involved in cellular
physiological and pathological processes, including cell
differentiation, proliferation, apoptosis and metabolism.
Currently, increased number of studies have shown that miRNA can
become a cancer biomarker and potential therapeutic target
(4).
After editing and maturation, miRNA can combine with
proteins into the RNA-induced silencing complex (RISC). In case
RISC does not fully match with the target mRNA gene, it can cut off
the miRNA translation and reduce the expression of target genes.
When RISC fully matches with the target mRNA gene, it can cause the
decomposition of mRNA. miRNA affects the target gene expression by
cutting off or decomposing target mRNA to regulate a series of
process, including cell differentiation and apoptosis (5,6).
The abnormal expression of miR-655 studied in this
test has been confirmed in a variety of tumor tissues and tumor
cells, including melanocytoma cells, lung adenocarcinoma cells and
pancreatic cancer cells (7,8). Studies have proved that miR-655 can
inhibit the epithelial-mesenchymal transition of tumor cells
(9), but there is no report on the
influence of miR-655 expression on the prognosis of ESCC at
present.
This investigation used the reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
technique to detect the expression of miR-655 in ESCC cell lines
and removed tissues in clinical surgery, upregulated the expression
of miR-655 in ESCC cells through mimic transfection to investigate
the influence on ESCC cell proliferation and metastasis ability.
The influence of miR-655 on the prognosis of ESCC was also studied
combined with clinical data analysis.
Materials and methods
Materials
The following were obtained: RPMI-1640 medium, fetal
calf serum (Gibco, Grand Island, NY, USA); RNA extraction kit
(Invitrogen Life Technologies, Carlsbad, CA, USA); RT-qPCR kit
(Takara Bio, Dalian, China); Transwell chamber (Corning Life
Sciences, Oneonta, NY, USA); Lipofectamine® 2000,
primer, miR-655 mimics (Invitrogen Life Technologies).
The human esophageal cancer cell lines, KYSE410 and
EC9706, were purchased from the Cell Bank of the Chinese Academy of
Sciences (Shanghai, China), and normal esophageal epithelial cells,
obtained via the isolation of the primary cells in 72 h culture,
were from the normal cancer-adjacent tissues. A total of 63 cases
of ESCC tissues and corresponding normal cancer-adjacent tissues
used in the clinical research were from the tissue specimens after
the esophageal cancer operation in Xuzhou Cancer Hospital (Xuzhou,
China). All tissue specimens were diagnosed as squamous cell
carcinoma by pathology analysis.
Cell culture
Two esophageal cancer cell lines, KYSE410 and
EC9706, were cultured in RPMI-1640 culture solution (containing 10%
fetal bovine serum, FBS) in 5% CO2 and incubated at
37°C, followed by passage once every 48–72 h. Cells in the
exponential phase were taken for the experiment.
Detection of miR-655 expression in
cells and specimens using RT-qPCR method
The total RNA was extracted from each group of cells
and tissues using the TRIzol method; 1 µg RNA was taken from the
total RNA obtained in each group, and the reverse transcription was
performed according to the operation method of the kit
specifications. Then miR-655 primer was added, and the expression
of miR-655 was detected using a two-step method for qPCR; U6 RNA
was selected as the internal reference. The specific operation was
in accordance with kit specifications. miR-655 primer sequence and
U6 primer sequence are shown in Table
I; the reaction conditions were as follows: 50°C for 2 min,
95°C for 1 min, 95°C for 5 sec, anneal and extension under 60°C for
34 sec, 40 amplification cycles.
 | Table I.Hsa-miR-655 mimics fragment sequences
and NC sequences. |
Table I.
Hsa-miR-655 mimics fragment sequences
and NC sequences.
| Gene | Primer sequences |
|---|
| miR-655 | F:
5′-TCCGAATAATACATGGTTAA-3′ |
|
| R:
5′-GTGCAGGGTCCGAGGT-3′ |
| U6 | F:
5′-TCCGATCGTGAAGCGTTC-3′ |
|
| R:
5′-GTGCAGGGTCCGAGGT-3′ |
| hsa-miR-655
mimics |
5′-AUAAUACAUGGUUAACCUCUUU-3′ |
| NC |
5′-UAUCUCCGAACGUGUCACGU-3′ |
Transfection of miR-655 mimics
Hsa-miR-655 mimics fragment and negative control
(NC) sequences are shown in Table I.
The transfection method was as follows: Cells were cultured using
6-well culture plate with the transfection of 2 µg per well; the
transfection was performed in accordance with the specification of
Lipofectamine® 2000; the experiment included the normal
group, miR-NC group and hsa-miR-655 mimics group.
Influence of miR-655 mimics
transfection on expression of miR-655 and cell proliferation
ability
At 48 h after transfection according to the above
method, the expression of miR-655 in cells in each group was
detected according to the methods previously specified. The cells
were transfected according to the above-mentioned method, digested
with trypsin after 48 h, and prepared into the single-cell
suspension using medium; then the suspension was added to the
96-well culture plate with 104 cells per well. The
experiment included the NC group and hsa-miR-655 mimics group. A
total of 20 µl MTT solution (concentration of 5 µg/µl) was added at
24 h. Cells were incubated in the dark for 4 h, and 150 µl DMSO was
added, followed by oscillation on the oscillator; the optical
density value in 490 nm wavelength was measured, then the cell
activity value was calculated.
Influence of miR-655 mimics
transfection on ESCC cell invasion ability
Transwell chamber was used, and the experiment
included NC group and hsa-miR-655 mimics group; after the chamber
was handled according to the operation method in the kit
specifications, the transfected cells were prepared into the
single-cell suspension containing 4×105/ml cells; 100 µl
cell suspension was added uniformly into the upper chamber, while
500 µl medium containing 30% FBS was added into the lower chamber.
After 48 h, specimens were fixed and dyed, followed by microscopic
observation and imaging analysis.
Relationship between the expression of
miR-655 in ESCC and prognosis
The inspection results in 1.3 were analyzed, and the
patients were divided into the miR-655 low-expression group and
miR-655 high-expression group according to whether the decrease of
miR-655 expression in ESCC tissues was more than doubled. There was
no statistical significance in the age, gender, smoking history and
other factors between the groups (P>0.05). According to the
follow-up results, the progression-free survival (PFS) of the
groups was calculated.
Statistical analysis
Data in this study are presented as mean ± standard
deviation; in data processing, SPSS 17.0 (SPSS, Inc., Chicago, IL,
USA) was used for one-way ANOVA. P≤0.05 indicates the difference
with statistical significance.
Results
Detection of miR-655 expression in
cells and specimens using RT-qPCR method
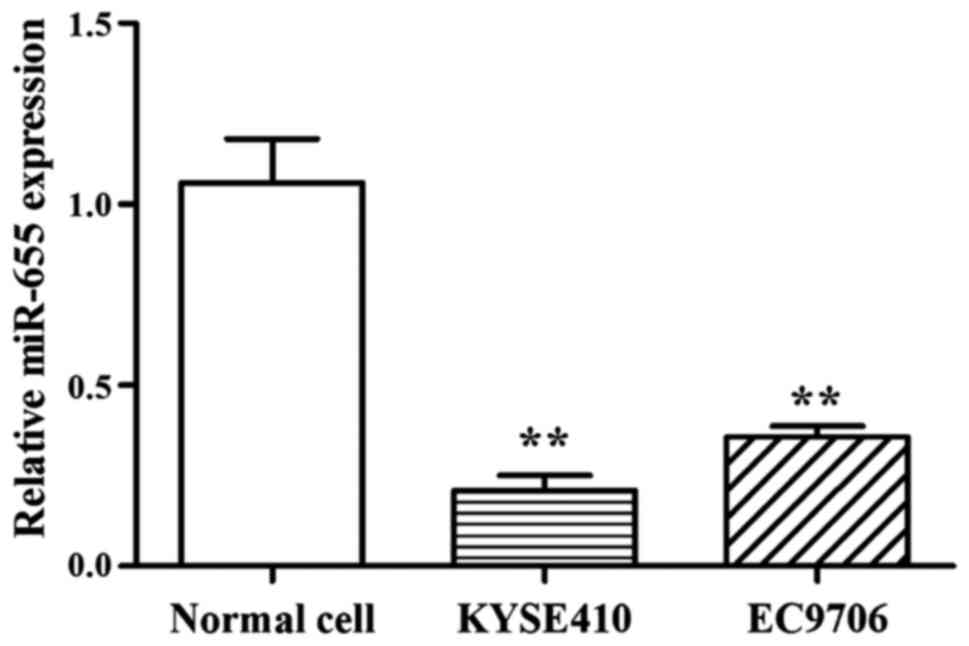
Results showed that compared with normal cells, the
expression of miR-655 was low in 2 ESCC cell lines, and the
relative expression of miR-655 was 0.21±0.04 (KYSE410) and
0.357±0.02 (EC9706) (Fig. 1).
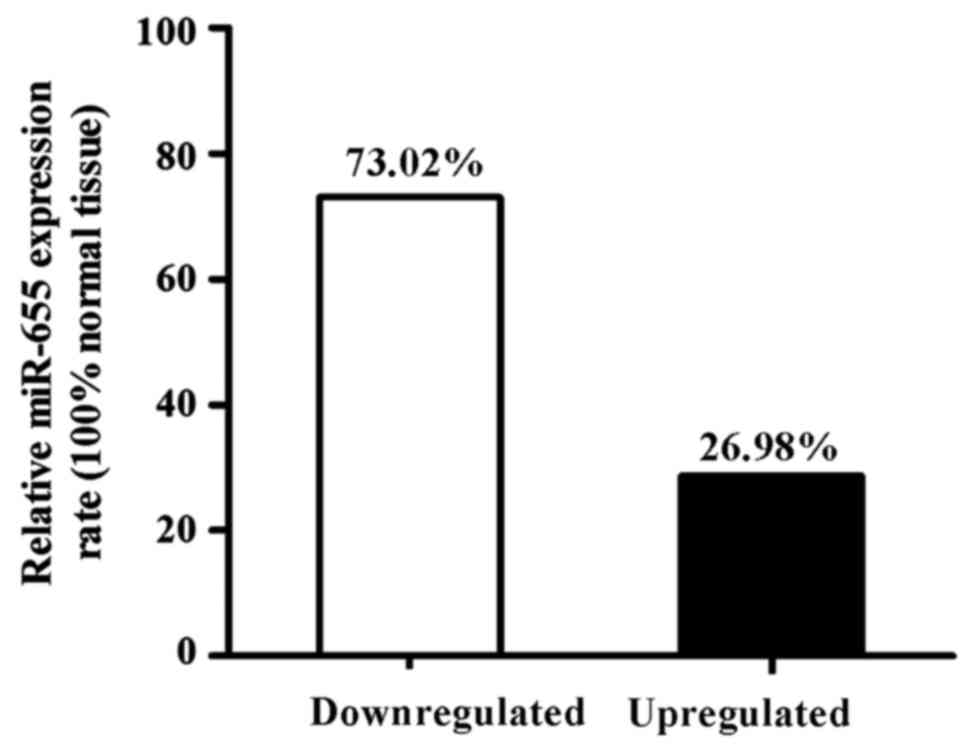
Among 63 ESCC specimens, the downregulated
expression of miR-655 relative to the normal cancer-adjacent
esophageal tissue was found in 46 patients (73.02%), and the
upregulated expression of miR-655 was found in 17 patients (26.98%)
(Fig. 2).
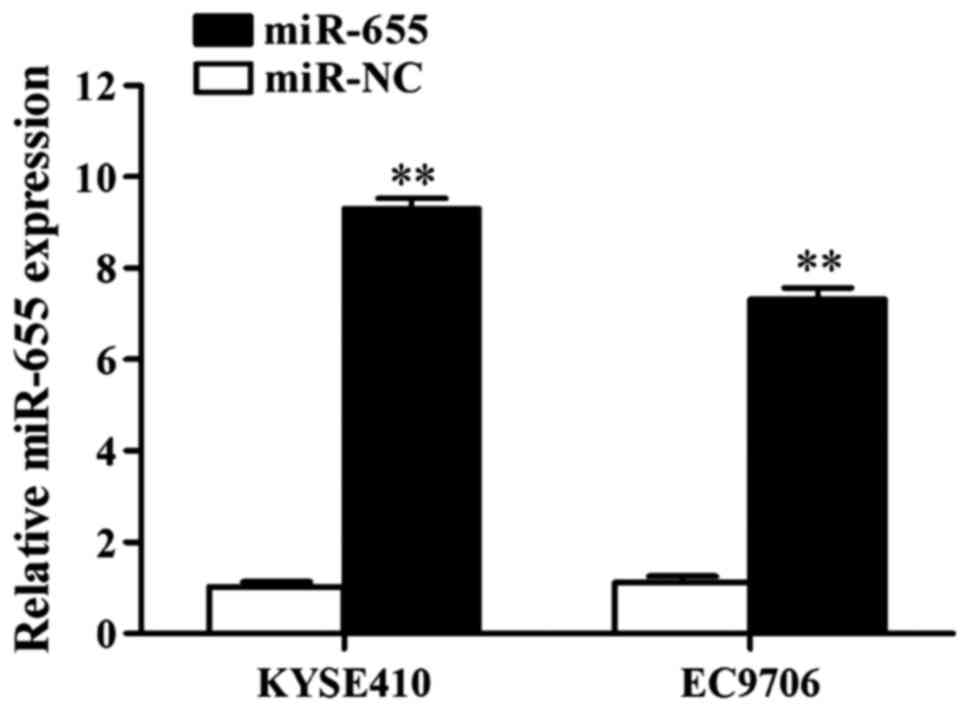
Influence of transfection of miR-655
mimics on miR-655 expression in ESCC cells
The influence of miR-655 mimics on miR-655
expression in ESCC cells was detected using RT-qPCR, and the
results showed that miR-655 mimics increased the expression of
miR-655 in KYSE410 cells 11.88-fold and in EC9706 cells 9.05-fold
(Fig. 3).
Influence of transfection of miR-655
mimics on ESCC cell proliferation ability
The influence of miR-655 on ESCC cell proliferation
was detected using MTT method. Results showed that after the
transfection of miR-655 mimics, the proliferation of KYSE410 and
EC9706 was inhibited 47.33 and 54.92%, respectively, and the
differences had statistical significance (P<0.01; Table II).
 | Table II.Influence of transfection of miR-655
mimics on ESCC cell proliferation ability. |
Table II.
Influence of transfection of miR-655
mimics on ESCC cell proliferation ability.
|
| Cell survival rate (%
miR-NC) |
|---|
|
|
|
|---|
| Group | KYSE410 | EC9706 |
|---|
| miR-NC | 100.75±4.41 | 101.28±3.52 |
| miR-655 |
47.33±3.76a |
54.92±2.47a |
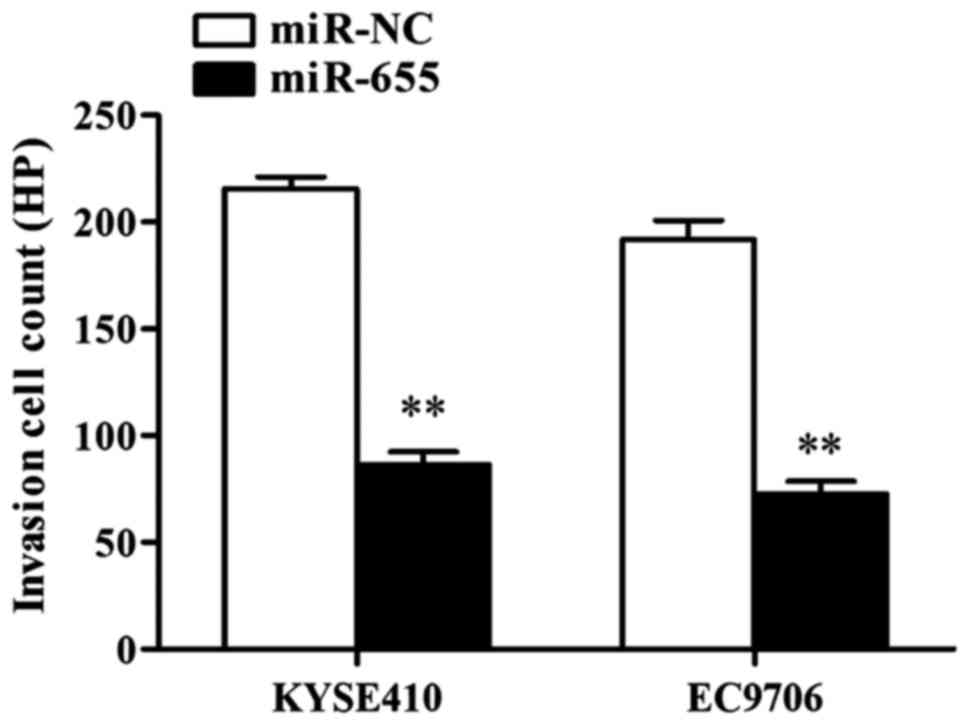
Influence of transfection of miR-655
mimics on ESCC cell invasion ability
The influence of miR-655 on ESCC cell invasion was
detected through Transwell cell invasion assay, and the results
showed that after KYSE410 was transfected with miR-655 mimics, the
number of cells crossing it was 215±7 cells/HP in NC group and 66±5
cells/HP in miR-655 transfection group, and the differences had
statistical significance (P<0.01); after EC9706 was transfected
with miR-655 mimics, the number of cells crossing it was 191±6
cells/HP in NC group and only 72±7 cells/HP in miR-655 transfection
group, and the differences had statistical significance
(P<0.01). The results showed that miR-655 effectively inhibited
the invasion ability of ESCC cells (Fig.
4).
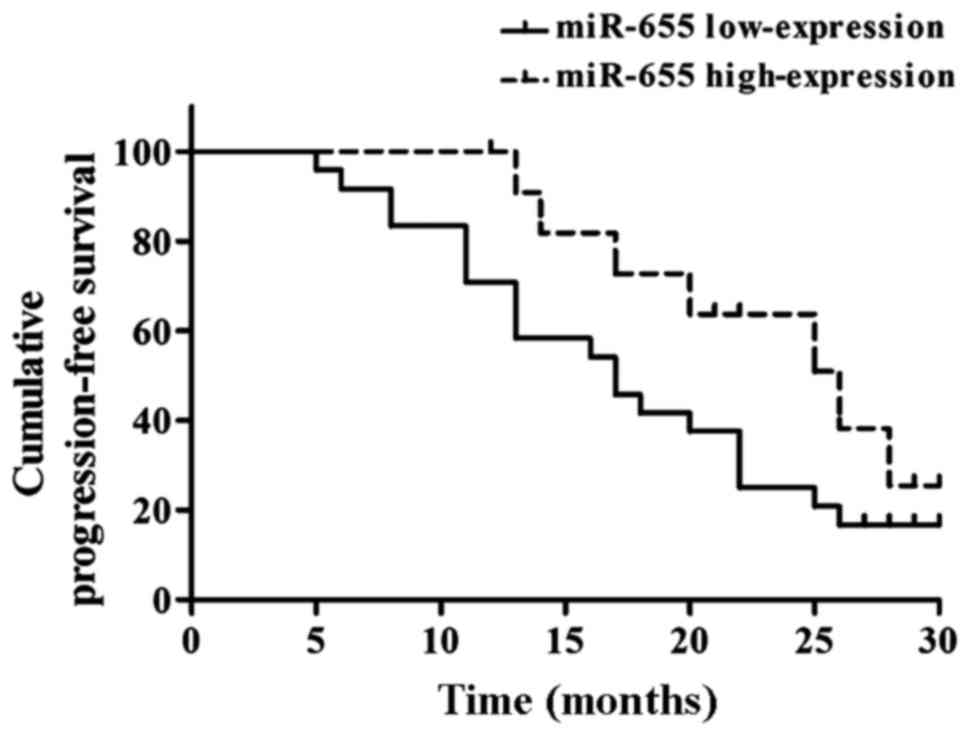
Relationship between miR-655
expression in ESCC and prognosis
Kaplan-Meier analysis suggested that the mean PFS
was 17.20±1.38 months in the miR-31 low-expression group and
26.50±1.23 months in the miR-655 high-expression group, and the
differences had statistical significance (Fig. 5).
Discussion
According to literature reports, both morbidity and
mortality of esophageal cancer are very high in China, and ranks as
fourth in malignant tumors, posing a great threat to people's
health and family life (10,11).
Studies have found that miRNA can regulate the
expression of tumor-related genes with the characteristics of
oncogenes or cancer suppressor genes. Studied have also found that
miRNA has a significant role in tumor inhibition during the process
of tumor generation (12). Some
reports also show that the expression of miR-21 is high in most
tumors and it acts as an oncogene (13). A large amount of data have proved that
the expression of miRNA is abnormal in a variety of tumors and this
is correlated to the diagnosis and prognosis of the tumor (14).
miRNA has high tissue specificity, which can be used
as both tumor suppressor gene and oncogene (15,16). miRNA
has a variety of biological characteristics, which can act as an
index of early tumor diagnosis and a predictive marker of prognosis
(5,17). miRNA is involved in tumor generation
and development. The expression of miRNA in tumor cells and normal
tissues is significantly different (1,18,19).
This study found that the expression of miR-655 in
ESCC cell lines was decreased significantly compared with that in
normal cells. Among 63 ESCC specimens, the downregulated expression
of miR-655 relative to the normal cancer-adjacent esophageal tissue
was found in 73.02% of specimens, and the upregulated expression of
miR-655 was found in 26.98% of specimens. In order to study the
influence of miR-655 on ESCC cells, an experimental study was
further performed and the expression of miR-655 in cells was
increased through the transfection. The results showed that the
high-expression of miR-655 could inhibit the proliferation and
invasion ability of ESCC cells. These results suggested that
miR-655 has the effect of inhibiting tumor in ESCC cells. In
addition, the clinical data showed that the median survival time of
patients with high-expression of miR-655 was longer than those with
low-expression of miR-655. Therefore, transferring miR-655 into
tumor cells can potentially prevent tumor generation to optimize
patient prognosis. miR-655 may be a kind of new biomarker to
diagnose and predict the prognosis of ESCC.
References
|
1
|
Mathé EA, Nguyen GH, Bowman ED, Zhao Y,
Budhu A, Schetter AJ, Braun R, Reimers M, Kumamoto K, Hughes D, et
al: MicroRNA expression in squamous cell carcinoma and
adenocarcinoma of the esophagus: Associations with survival. Clin
Cancer Res. 15:6192–6200. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Fareed KR, Kaye P, Soomro IN, Ilyas M,
Martin S, Parsons SL and Madhusudan S: Biomarkers of response to
therapy in oesophago-gastric cancer. Gut. 58:127–143. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Farazi TA, Spitzer JI, Morozov P and
Tuschl T: miRNAs in human cancer. J Pathol. 223:102–115. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Rosenfeld N, Aharonov R, Meiri E,
Rosenwald S, Spector Y, Zepeniuk M, Benjamin H, Shabes N, Tabak S,
Levy A, et al: MicroRNAs accurately identify cancer tissue origin.
Nat Biotechnol. 26:462–469. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Hobert O: Gene regulation by transcription
factors and microRNAs. Science. 319:1785–1786. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wang Y, Zang W, Du Y, Ma Y, Li M, Li P,
Chen X, Wang T, Dong Z and Zhao G: Mir-655 up-regulation suppresses
cell invasion by targeting pituitary tumor-transforming gene-1 in
esophageal squamous cell carcinoma. J Transl Med. 11:3012013.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kitamura K, Seike M, Okano T, Matsuda K,
Miyanaga A, Mizutani H, Noro R, Minegishi Y, Kubota K and Gemma A:
MiR-134/487b/655 cluster regulates TGF-β-induced
epithelial-mesenchymal transition and drug resistance to gefitinib
by targeting MAGI2 in lung adenocarcinoma cells. Mol Cancer Ther.
13:444–453. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Harazono Y, Muramatsu T, Endo H, Uzawa N,
Kawano T, Harada K, Inazawa J and Kozaki K: miR-655 is an
EMT-suppressive microRNA targeting ZEB1 and TGFBR2. PLoS One.
8:e627572013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Parkin DM, Bray F, Ferlay J and Pisani P:
Global cancer statistics, 2002. CA Cancer J Clin. 55:74–108. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chen W, Zheng R, Zeng H, Zhang S and He J:
Annual report on status of cancer in China, 2011. Chin J Cancer
Res. 27:2–12. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Calin GA, Dumitru CD, Shimizu M, Bichi R,
Zupo S, Noch E, Aldler H, Rattan S, Keating M, Rai K, et al:
Frequent deletions and down-regulation of micro- RNA genes miR15
and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad
Sci USA. 99:pp. 15524–15529. 2002; View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Pan X, Wang ZX and Wang R: MicroRNA-21: A
novel therapeutic target in human cancer. Cancer Biol Ther.
10:1224–1232. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Calin GA and Croce CM: MicroRNA signatures
in human cancers. Nat Rev Cancer. 6:857–866. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Sempere LF, Christensen M, Silahtaroglu A,
Bak M, Heath CV, Schwartz G, Wells W, Kauppinen S and Cole CN:
Altered MicroRNA expression confined to specific epithelial cell
subpopulations in breast cancer. Cancer Res. 67:11612–11620. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Medina PP, Nolde M and Slack FJ: OncomiR
addiction in an in vivo model of microRNA-21-induced pre-B-cell
lymphoma. Nature. 467:86–90. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Schaefer A, Jung M, Mollenkopf HJ, Wagner
I, Stephan C, Jentzmik F, Miller K, Lein M, Kristiansen G and Jung
K: Diagnostic and prognostic implications of microRNA profiling in
prostate carcinoma. Int J Cancer. 126:1166–1176. 2010.PubMed/NCBI
|
|
18
|
Davis E, Caiment F, Tordoir X, Cavaillé J,
Ferguson-Smith A, Cockett N, Georges M and Charlier C:
RNAi-mediated allelic trans-interaction at the imprinted Rtl1/Peg11
locus. Curr Biol. 15:743–749. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Castilla MÁ, Moreno-Bueno G, Romero-Pérez
L, Van De Vijver K, Biscuola M, López-García MÁ, Prat J,
Matías-Guiu X, Cano A, Oliva E, et al: Micro-RNA signature of the
epithelial-mesenchymal transition in endometrial carcinosarcoma. J
Pathol. 223:72–80. 2011. View Article : Google Scholar : PubMed/NCBI
|