Introduction
Prostate cancer is one of the most common malignant
tumors in the urinary and reproductive system (1–3). Prostate
cancer is the second most common cancer globally and has the sixth
highest mortality rate (4). In
previous years, microRNA (miRNA) regulation and its role in the
occurrence and development of prostate cancer has become a focus of
study.
miRNAs are 21–25 nucleotide non-coding,
single-stranded RNA, which regulate gene expression at the
transcriptional and translational level. miRNAs may regulate tumor
occurrence, development and prognosis through the oncogenes or
tumor suppressor genes (5). Due to
the different expression of miRNAs in normal tissues and malignant
tissues, the expression profile of miRNAs in different tissues may
be a potential tool for the diagnosis, targeted therapy and
prognosis of tumors. In 2007, Porkka et al (6) used gene chip technology for the first
time to report the different expression pattern of miRNAs in benign
and malignant prostate tumors. Ambs et al (7) studied the differences in miRNA and mRNA
in 60 cases of prostate cancer and normal prostate tissue and
revealed that miRNA is associated with the development of prostate
cancer, and miRNA controls tumor growth by regulating the
expression of tumor-associated genes to promote prostate cancer
occurrence and development. Chen et al (8) and other comparative studies (9,10) of the
expression of miRNA in the plasma of patients with prostate cancer
and benign prostatic hyperplasia revealed that the expression of
miR-622 and miR-1285 significantly increased, while the expression
of let-7e, Let-7c and miR-30c significantly decreased. Therefore,
the present study hypothesized that the differential expression of
the 5 miRNAs in the plasma may be used as a biochemical marker and,
combined with the prostate-specific antigen test, can be used in
the diagnosis of prostate cancer. Sylvestre et al (11) and other studies (12) demonstrated that miR-20a is
overexpressed in prostate cancer, and overexpression of miR-20a in
PC-3 cells exhibits an anti-apoptotic effect. Qin et al
(13) reported that miR-24 regulates
the expression of the target gene fas associated factor 1, and
induces the apoptosis of hormone refractory prostate cancer cells.
Bin et al confirmed that miR-146a (14) inhibits the progression of prostate
cancer tumors through the epidermal growth factor receptor
signaling pathway.
Previous studies have demonstrated that miR-30c is
expressed in numerous types of malignant tumors, including breast,
ovary, stomach and prostate cancers. Lee et al (15) previously reported that mir-30c had
significantly lower expression in ovarian cancer tissue compared
with normal ovarian tissue, and additional multivariate analysis
revealed that patients with ovarian cancer mir-30c high expression
had longer survival and overall survival rate. Zhou et al
(16) revealed that miR-30c is a
tumor suppressor gene that can inhibit the growth of endometrial
cancer by targeting the expression of metastasis-associated 1. In
lung cancers, Zhong et al demonstrated that low expression
of miR-30c promotes invasion by inducing epithelial mesenchymal
transition (17). Wu et al
(18) identified that miR-30c
overexpression inhibits the migration and invasion of the
hepatocellular carcinoma cell lines SMMC-7721 and HepG2. Therefore,
overexpression of miR-30c may inhibit the proliferation, migration
and invasion of tumor cells. However, Rodríguez-González et
al (19) reported that high
expression of miR-30c may be used as an independent predictor of
the efficacy of endocrine therapy for breast cancer and tumor
progression free survival. Dobson et al (20) demonstrated that overexpression of
miR-30c in the breast cancer MDA-MB-231 cell line promotes the
invasive metastatic phenotype. Therefore, the role of miR-30c in
different tumors remains controversial.
Previous studies have reported that plasma mir-30c
exhibited low expression in prostate cancer tissue (6,8). However,
the mechanism of miR-30c involvement in the development and
progression of prostate cancer remains unclear, and the target gene
is not clear. The present study aimed to examine the underlying
mechanism of the association between miR-30c and prostate cancer
growth in order to provide additional evidence to facilitate the
improvement of therapeutic strategies for this disease.
Materials and methods
Cell culture
LNCaP, DU145, PC-3 and RWPE-1 (American Type Culture
Collection, Manassas, VA, USA) were cultured in Roswell Park
Memorial Institute (RPMI)-1640 medium with 10% fetal bovine serum
(FBS; Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA)
and penicillin (100 U/ml). The cells were cultured at 37°C with 5%
CO2.
Plasmid construction and
transfection
A miR-30c expression vector was constructed by
cloning the fragment, amplified from human genomic DNA by
polymerase chain reaction (PCR) with the primers,
hmiR30c-EcoRI (forward,
5′-CCGGAATTCAACATAGTGTGGGGATGGGGT-3′) and hmiR30c-BamHI
(reverse, 5′-CGCGGATCCAGGTTAATGGGAAACAGGGCT-3′) into the
EcoRI and BamHI sites of the pLV-mCherry(2A)puro
vector. The pLV-mCherry(2A)puro vector and packaging vector were
co-transfected into HEK293T cells (American Type Culture
Collection) to obtain high titer virus particles with replication
defects. For lentiviral infection, cells were plated at a
concentration of 1×105 cells/well onto 6-well culture
plates and then infected with indicated lentiviruses in the
presence of 8 µg/ml polybrene. Following infection for 72 h,
puromycin was added to the medium at a concentration of 0.5 µg/ml
for PC-3 cells and cell populations were selected for 2 weeks for
following experiments.
Isolation of total RNA and
quantitative PCR (qPCR)
Total RNA was extracted from collected cells using
TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.) and
mRNA was then reverse transcribed to cDNA (Promega Corporation,
Madison, WI, USA). U6 small nuclear RNA was used for normalization.
The PCR reactions were performed with the following primers:
hsa-miR-30c forward, 5′-CCCGCTGTAAACATCCTACACTC-3′ and reverse,
5′-GTGCAGGGTCCGAGGT-3′; and U6 forward, 5′-CGCTTCGGCAGCACATATAC-3′
and reverse, 5′-CAGGGGCCATGCTAATCTT-3′. qPCR was performed using
the ABI 7900 Fast Real-Time PCR system (Applied Biosystems; Thermo
Fisher Scientific, Inc.).
Cell cycle analysis
The cell cycle was assessed at 24 h following
transfection by propidium iodide staining, and measured with a FACS
Calibur flow cytometer (BD Biosciences, San Jose, CA, USA). MTT
PC-3 cells were trypsinized and seeded onto 96-well plates at a
density of 3×103 cells/well for culture. Cell
proliferation was measured using the MTT assay 24, 48 and 72 h
after seeding. Briefly, 10 µl of MTT solution (5 mg/ml) was added
to each well, and the cells were incubated for 3 h at 37°C. The
medium was then aspirated, and 150 µl of dimethyl sulfoxide was
added. The optical density was measured at 570 nm with a microplate
reader (Bio-Rad Laboratories, Inc., Hercules, CA, USA). Three
independent experiments were performed in quadruplicate. The cell
growth inhibition rate (%)=(1-value of experimental group/negative
control group) ×100.
Colony formation assay
The two cells types were harvested and 1,000 cells
were inoculated in each pore of a 6-well plate. The medium was
changed every 2 days. After 14 days, cells were rinsed with 1X PBS
and then fixed with methanol for 10 min. Subsequently, cells were
stained with 0.1% crystal violet (Sigma-Aldrich; Merck KGaA,
Darmstadt, Germany) for >1 h.
Wound healing assay
The cells were seeded onto 6-well plates and
cultured with RPMI-1640 medium. Following 24 h, the cells were
wounded with a 200 µl pipette tip. Serum-free RPMI-1640 medium was
added and wound closure was observed for 30 h using an XSP-4C
microscope at a magnification of ×200 (Shanghai Changfang Optical
Instrument Co. Ltd., Shanghai, China).
Transwell assay
Cell motility was measured using an 8 µm pore
polycarbonate membrane Boyden chamber insert in a Transwell
apparatus (EMD Millipore, Billerica, MA, USA). The transfected
cells were treated with 0.2% trypsin/EDTA solution and washed once
with serum-containing RPMI-1640 medium. A total of
1.5×105 cells in 0.2 ml serum-free RPMI-1640 medium were
seeded onto a Transwell apparatus. RPMI-1640 medium containing 10%
FBS (600 µl) was added to the lower chamber. An invasion assay was
conducted following the aforementioned procedure, with the
exception that the filters of the Transwell chambers were coated
with 45 µg Matrigel (BD Biosciences). Following incubation of the
cells for 24, 48 and 72 h at 37°C in a 5% CO2 incubator,
the cells on the top surface of the insert were removed by wiping
with a cotton swab. The cells that invaded to the bottom surface of
the insert were fixed in the 100% precooling methanol for 10 min,
stained with 0.1% crystal violet for 30 min, rinsed in PBS and then
subjected to microscopic inspection (XSP-4C microscope;
magnification, ×200). The values for invasion were obtained by
counting three fields per membrane and were represented as the
average of the three independent experiments.
Western blot analysis
The total proteins were prepared from the
established cells. The proteins were fractionated by SDS-PAGE,
transferred to a polyvinylidene fluoride membrane (EMD Millipore),
blocked in 5% dry milk at room temperature for 1 h and
immunostained with antibodies at 4°C overnight using anti-KRAS
(dilution, 1:1,000; #sc-30; Santa Cruz Biotechnology, Inc., Dallas,
TX, USA) and anti-GAPDH (dilution, 1:5,000; #sc-47724; Santa Cruz
Biotechnology, Inc.). All results were visualized through an
electrochemiluminescence substrate western blotting detection
system (Pierce; Thermo Fisher Scientific, Inc.) and then exposed by
the Molecular Imager ChemiDoc XRS System (Bio-Rad Laboratories,
Inc.). The integrated density of the band was quantified by ImageJ
software v1.37 (Bio-Rad Laboratories, Inc.).
Statistical analysis
The 2−∆∆Cq method was used to analyze the
results of qPCR in all the experiments performed in the present
study (21). Statistical analysis was
performed using SPSS 10.0 (SPSS, Inc., Chicago, IL, USA) and
presented with GraphPad prism software v6.01 (GraphPad Software,
Inc., La Jolla, CA, USA). The results obtained from experiment
in vitro assays are presented as the mean ± standard
deviation from at least 3 independent experiments, and the data
were analyzed by the t-test. P<0.05 was considered to indicate a
statistically significant difference.
Results
Expression of miR-30c in the PC-3 cell
line is lower than other human prostate cell lines
It was reported that the expression level of miR-30c
in prostate cancer tissue and plasma was low (22). In order to study the function of
miR-30c in the prostate cell lines, the expression of miR-30c in
human prostate cell lines was tested by reverse transcription-qPCR.
PC-3 was revealed to possess significantly lower miR-30c expression
than the LNCaP, DU145 and RWPE-1 cell lines (P<0.001; Fig. 1). Therefore, based on this expression
pattern, the PC-3 cell line was selected to verify the effect of
miR-30c.
miR-30c regulates the cell cycle of
PC-3 cells in vitro
To examine the mechanism underlying the effect of
miR-30c on tumor growth in prostate cancer, the PC-3 cells were
transfected with control miRNA (NC) and miR-30c. The transfection
efficiency was validated by qPCR. The cell cycle assay performed by
fluorescence correlation spectroscopy demonstrated that
overexpression of miR-30c caused the accumulation of PC-3 cells in
the G2/M phase compared with the NC (P<0.05; Fig. 2). The results indicated that miR-30c
may inhibit growth of the PC-3 cell line in vitro.
miR-30c overexpression inhibits the
growth of the PC-3 cell line
To verify whether miR-30c regulates tumor growth in
prostatic cancer, the MTT test was used to study PC-3 cell growth,
which had been transfected with miR-30c NC or miR-30c. The
transfection efficiency was validated by qPCR. Overexpression of
miR-30c significantly inhibited the growth of PC-3 cells compared
with the control at 48 and 72 h (P<0.05; Fig. 3). The present results indicated that
miR-30c inhibits the growth of the PC-3 cell line in
vitro.
Role of miR-30c overexpression in the
PC-3 cell line colony formation ability
To assess whether the overexpression of miR-30c
expression affected the tumorigenic properties of the cells, the
colony formation assay was used (Fig.
4). Following inoculation for 14 days, miR-30c transfected
cells exhibited a significantly lower colony formation ability
compared with the control cells. Therefore, it was concluded that
miR-30c contributed to the regulation of PC-3 cell line growth.
miR-30c regulates the invasion of PC-3
cells in vitro
To examine whether miR-30c affects tumor cell
invasion, the PC-3 cells were transfected with miR-30c and NC. The
transfection efficiency was validated by qPCR. The wound healing
assay demonstrated that the overexpression of miR-30c was able to
suppress PC-3 cell healing (Fig. 5).
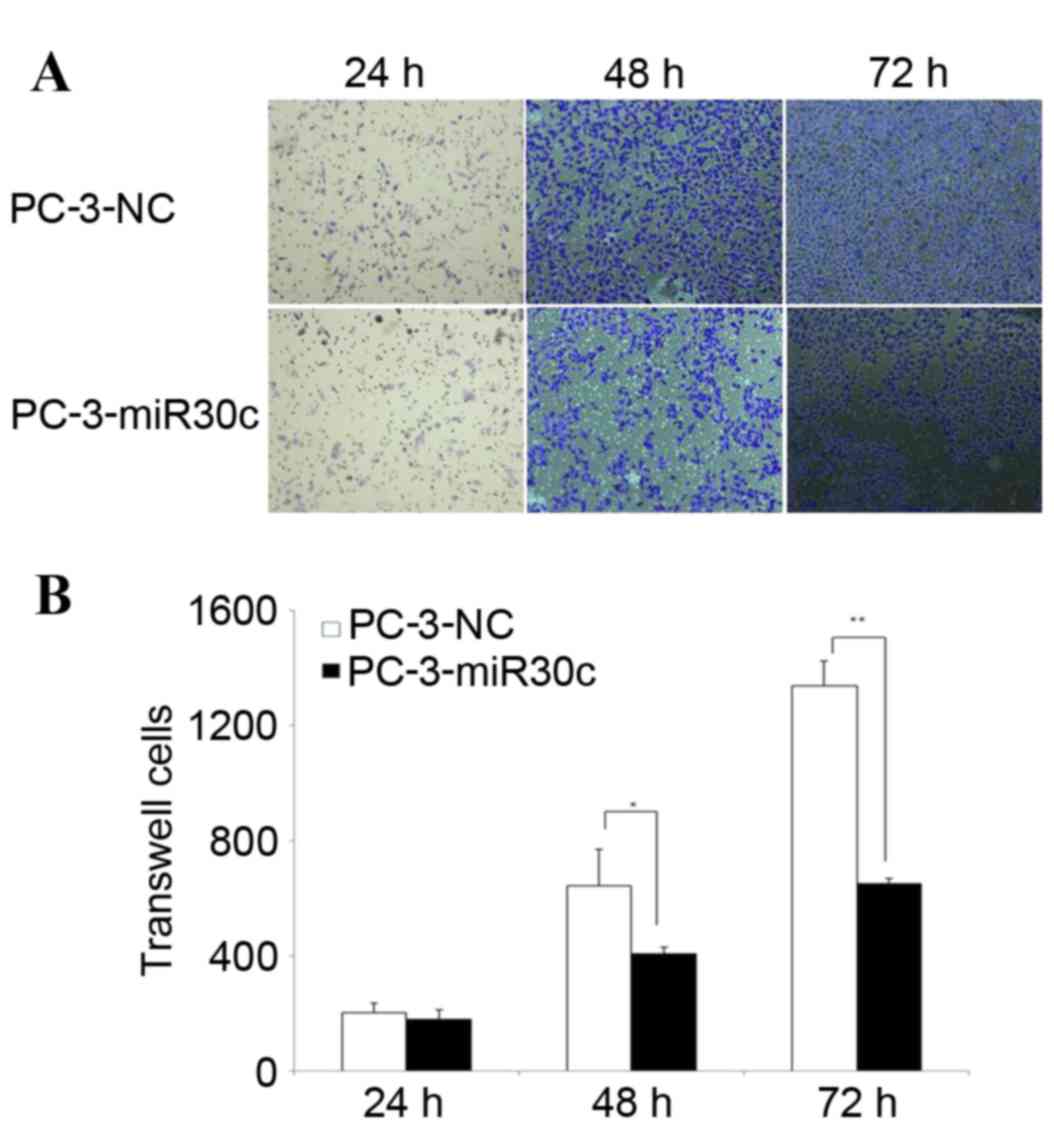
Furthermore, the Transwell assay demonstrated that overexpression
of miR-30c attenuated PC-3 cell invasion (Fig. 6). These results indicated that
overexpression of miR-30c may inhibit invasion of the PC-3 cell
line in vitro.
Overexpression of miR-30c is
associated with the downregulation of KRAS expression
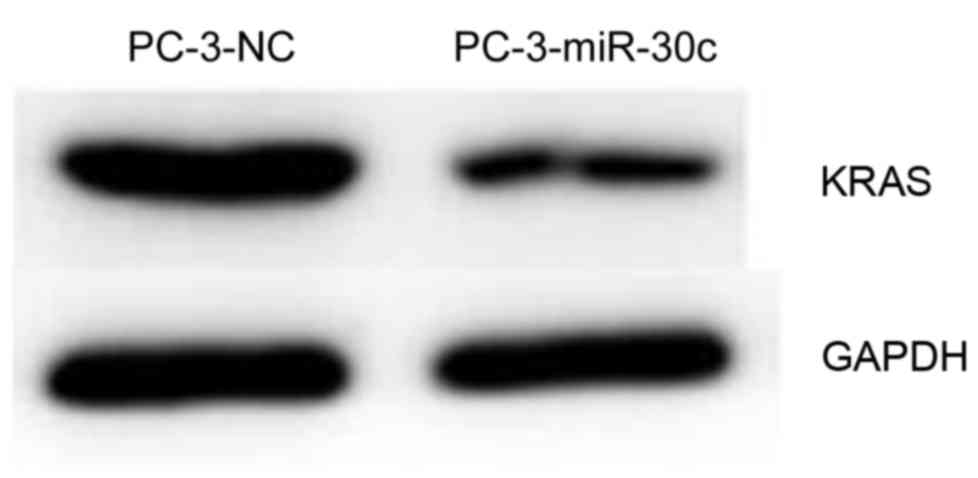
It was previously reported that KRAS is a target of
miR-30c in breast cancer (23). The
present study aimed to investigate whether miR-30c can control KRAS
in prostatic cancer. Using western blot analysis, overexpression of
miR-30c was revealed to significantly inhibit the expression of
KRAS protein compared with the control (Fig. 7). The present data indicated that
there may be a common underlying mechanism of tumor
progression.
Discussion
In the present study, it was demonstrated that
overexpression of miR-30c inhibits prostate cancer cell line
proliferation, migration and invasion, which is possibly involved
in downregulation of KRAS protein. Thus, the data provided support
a mechanism by which miR-30c serves a tumor suppressive role in
prostate cancer.
It was previously reported (6,8) that
plasma mir-30c exhibits low expression in prostate cancer tissue.
At present, however, the mechanism of miR-30c involvement in the
development and progression of prostate cancer remains unclear, and
the target gene is also uncertain. The present study aimed to
examine the underlying mechanism of the association between miR-30c
and prostate cancer growth, in order to provide additional evidence
to facilitate improvement of the therapeutic strategies for this
disease.
miRNA expression profiles are distinct among
different tumor types and often reflect the tissue of origin and
differentiation state of the tumor (24,25).
Previous studies have shown that miR-30c is overexpressed in breast
cancer (26,18). By contrast, miR-30c has low expression
in prostate cancer tissue and plasma. The role of miR-30c involved
in the development and progression of prostate cancer is not clear,
and the target gene is not clear. In the present study, it was
confirmed that miR-30c expression is lower in the PC-3 cell line
compared with other prostate cancer cell lines; therefore, the PC-3
cell line was used to study the function of miR-30c. To investigate
the role of miR-30c in prostate cancer, PC-3 cell lines
overexpressing miR-30c were used. Reduced tumor proliferation and
cell cycle block was identified in miR-30c overexpression PC-3
cells compared with negative control miRNA overexpression PC-3
cells, which indicated that the miR-30c suppresses tumor
metastasis.
It was previously reported that miR-30c can regulate
KRAS in breast cancer (23), and the
present study revealed that KRAS was downregulated in miR-30c
overexpression cells compared with control cells. This indicated
that KRAS may be a common target of miR-30c in different types of
tumors.
In conclusion, the present study demonstrated that
overexpression of miR-30c inhibits prostate cancer cell
proliferation, migration and invasion. miR-30c-mediated inhibition
of PC-3 invasive phenotypes may be caused by the downregulation of
KRAS protein.
Acknowledgements
The authors acknowledge financial support from the
Health and Family Planning Commission Foundation of Shanghai
Minhang District (grant no. 2013MW05) and the Shanghai Key Medical
Specialty Program (grant no. ZK2015B04).
References
|
1
|
Peyromaure EM, Mao K, Sun Y, Xia S, Jiang
N, Zhang S, Wang G, Liu Z and Debré B: A comparative study of
prostate cancer detection and management in China and in France.
Can J Urol. 16:4472–4477. 2009.PubMed/NCBI
|
|
2
|
Zeigler-Johnson CM, Rennert H, Mittal RD,
Jalloh M, Sachdeva R, Malkowicz SB, Mandhani A, Mittal B, Gueye SM
and Rebbeck TR: Evaluation of prostate cancer characteristics in
four populations worldwide. Can J Urol. 15:4056–4064.
2008.PubMed/NCBI
|
|
3
|
Torre LA, Siegel RL, Ward EM and Jemal A:
Global cancer incidence and mortality rates and Trends-An update.
Cancer Epidemiol Biomarkers Prev. 25:16–27. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Shi XB, Tepper CG and White RW deVere:
Cancerous miRNAs and their regulation. Cell Cycle. 7:1529–1538.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Porkka KP, Pfeiffer MJ, Waltering KK,
Vessella RL, Tammela TL and Visakorpi T: MicroRNA expression
profiling in prostate cancer. Cancer Res. 67:6130–6135. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ambs S, Prueitt RL, Yi M, Hudson RS, Howe
TM, Petrocca F, Wallace TA, Liu CG, Volinia S, Calin GA, et al:
Genomic profiling of microRNA and messenger RNA reveals deregulated
microRNA expression in prostate cancer. Cancer Res. 68:6162–6170.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Chen ZH, Zhang GL, Li HR, Luo JD, Li ZX,
Chen GM and Yang J: A panel of five circulating microRNAs as
potential biomarkers for prostate cancer. Prostate. 72:1443–1452.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kachakova D, Mitkova A, Popov E, Popov I,
Vlahova A, Dikov T, Christova S, Mitev V, Slavov C and Kaneva R:
Combinations of serum prostate-specific antigen and plasma
expression levels of let-7c, miR-30c, miR-141 and miR-375 as
potential better diagnostic biomarkers for prostate cancer. DNA
Cell Biol. 34:189–200. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Cochetti G, Poli G, Guelfi G, Boni A,
Egidi MG and Mearini E: Different levels of serum microRNAs in
prostate cancer and benign prostatic hyperplasia: Evaluation of
potential diagnostic and prognostic role. Onco Targets Ther.
9:7545–7553. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Sylvestre Y, De Guire V, Querido E,
Mukhopadhyay UK, Bourdeau V, Major F, Ferbeyre G and Chartrand P:
An E2F/miR-20a autoregulatory feedback loop. J Biol Chem.
282:2135–2143. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Dhar S, Kumar A, Rimando AM, Zhang X and
Levenson AS: Resveratrol and pterostilbene epigenetically restore
PTEN expression by targeting oncomiRs of the miR-17 family in
prostate cancer. Oncotarget. 6:27214–27226. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Qin W, Shi Y, Zhao B, Yao C, Jin L, Ma J
and Jin Y: miR-24 regulates apoptosis by targeting the open reading
frame (ORF) region of FAF1 in cancer cells. PLoS One. 5:e94292010.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Xu B, Wang N, Wang X, Tong N, Shao N, Tao
J, Li P, Niu X, Feng N, Zhang L, et al: MiR-146a suppresses tumor
growth and progression by targeting EGFR pathway and in a
p-ERK-dependent manner in castration-resistant prostate cancer.
Prostate. 72:1171–1178. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Lee H, Park CS, Deftereos G, Morihara J,
Stern JE, Hawes SE, Swisher E, Kiviat NB and Feng Q: MicroRNA
expression in ovarian carcinoma and its correlation with
clinicopathological features. World J Surg Oncol. 10:1742012.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhou H, Xu X, Xun Q, Yu D, Ling J, Guo F,
Yan Y, Shi J and Hu Y: microRNA-30c negatively regulates
endometrial cancer cells by targeting metastasis-associated gene-1.
Oncol Rep. 27:807–812. 2012.PubMed/NCBI
|
|
17
|
Zhong Z, Xia Y, Wang P, Liu B and Chen Y:
Low expression of microRNA-30c promotes invasion by inducing
epithelial mesenchymal transition in non-small cell lung cancer.
Mol Med Rep. 10:2575–2579. 2014.PubMed/NCBI
|
|
18
|
Wu W, Zhang X, Liao Y, Zhang W, Cheng H,
Deng Z, Shen J, Yuan Q, Zhang Y and Shen W: miR-30c negatively
regulates the migration and invasion by targeting the immediate
early response protein 2 in SMMC-7721 and HepG2 cells. Am J Cancer
Res. 5:1435–1446. 2015.PubMed/NCBI
|
|
19
|
Rodríguez-González FG, Sieuwerts AM, Smid
M, Look MP, Meijer-van Gelder ME, De Weerd V, Sleijfer S, Martens
JW and Foekens JA: MicroRNA-30c expression level is an independent
predictor of clinical benefit of endocrine therapy in advanced
estrogen receptor positive breast cancer. Breast Cancer Res Treat.
127:43–51. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Dobson JR, Taipaleenmäki H, Hu YJ, Hong D,
van Wijnen AJ, Stein JL, Stein GS, Lian JB and Pratap J:
hsa-mir-30c promotes the invasive phenotype of metastatic breast
cancer cells by targeting NOV/CCN3. Cancer Cell Int. 14:732014.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Chen ZH, Zhang GL, Li HR, Luo JD, Li ZX,
Chen GM and Yang J: A panel of five circulating microRNAs as
potential biomarkers for prostate cancer. Prostate. 72:1443–1452.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Tanic M, Yanowsky K, Rodriguez-Antona C,
Andrés R, Márquez-Rodas I, Osorio A, Benitez J and Martinez-Delgado
B: Deregulated miRNAs in hereditary breast cancer revealed a role
for miR-30c in regulating KRAS oncogene. PLoS One. 7:e388472012.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Jay C, Nemunaitis J, Chen P, Fulgham P and
Tong AW: miRNA profiling for diagnosis and prognosis of human
cancer. DNA Cell Biol. 26:293–300. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Lu J, Getz G, Miska EA, Alvarez-Saavedra
E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA,
et al: MicroRNA expression profiles classify human cancers. Nature.
435:834–838. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Yen MC, Shih YC, Hsu YL, Lin ES, Lin YS,
Tsai EM, Ho YW, Hou MF and Kuo PL: Isolinderalactone enhances the
inhibition of SOCS3 on STAT3 activity by decreasing miR-30c in
breast cancer. Oncol Rep. 35:1356–1364. 2016.PubMed/NCBI
|