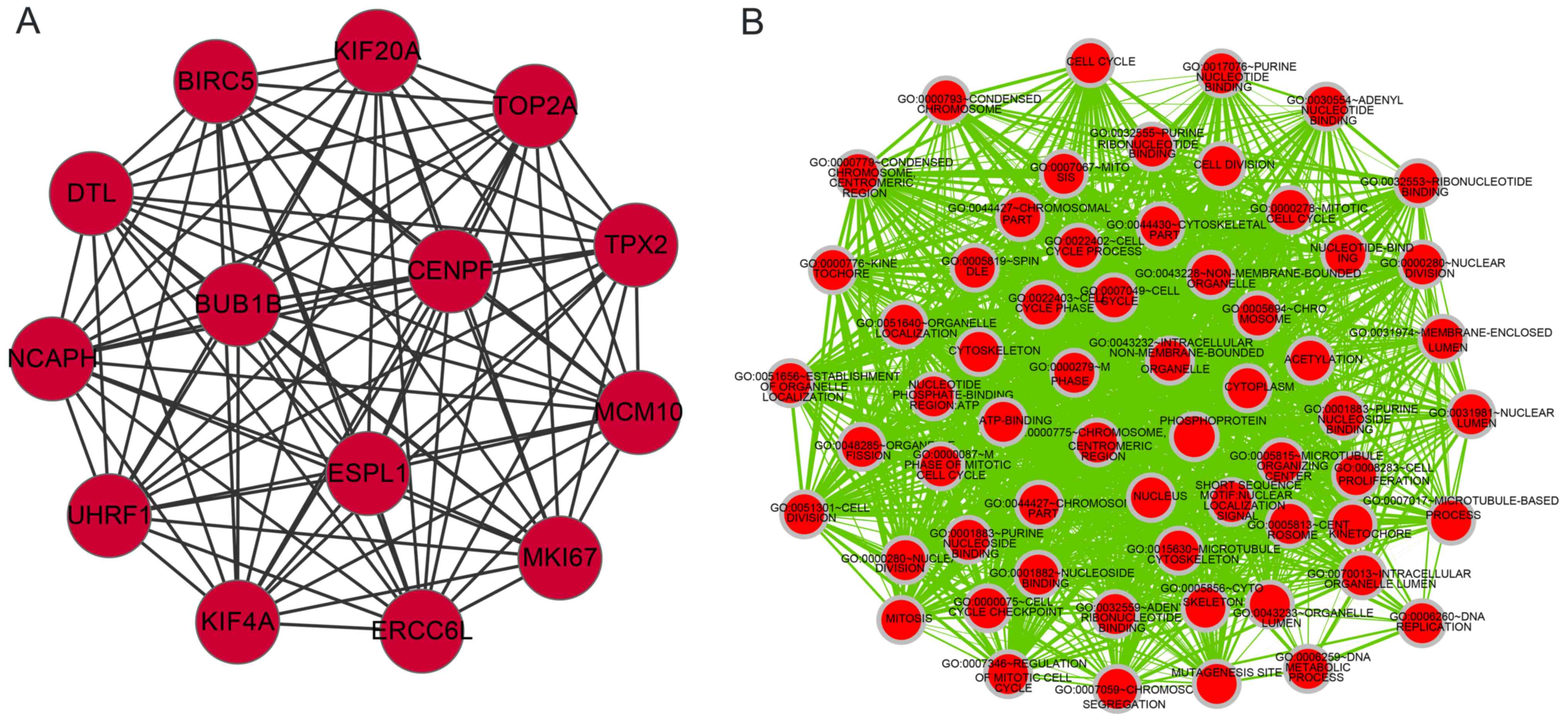
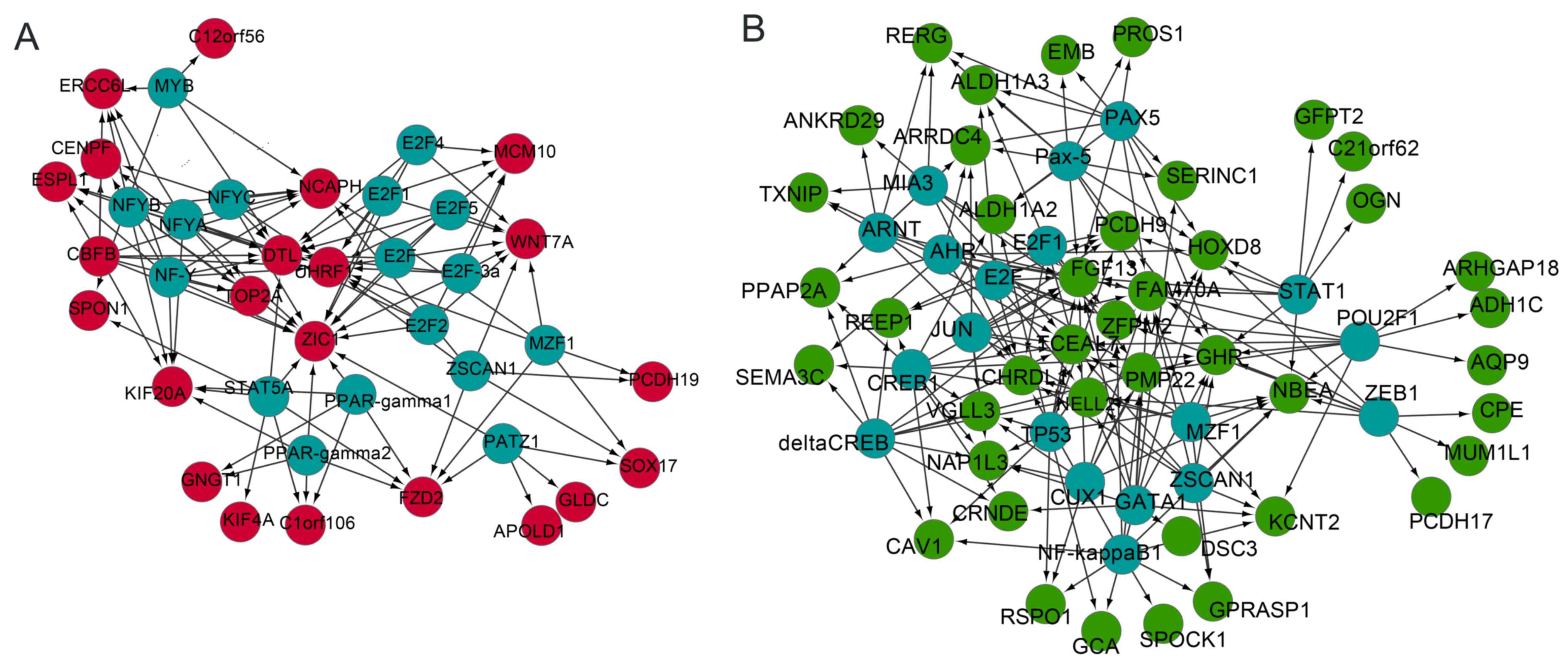
|
1
|
Jemal A, Siegel R, Xu J and Ward E: Cancer
statistics, 2010. CA Cancer J Clin. 60:277–300. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Marchetti C, Pisano C, Facchini G, Bruni
GS, Magazzino FP, Losito S and Pignata S: First-line treatment of
advanced ovarian cancer: Current research and perspectives. Expert
Rev Anticancer Ther. 10:47–60. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Sansone P and Bromberg J: Targeting the
interleukin-6/Jak/stat pathway in human malignancies. J Clin Oncol.
30:1005–1014. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Colomiere M, Ward AC, Riley C, Trenerry
MK, Cameron-Smith D, Findlay J, Ackland L and Ahmed N: Cross talk
of signals between EGFR and IL-6R through JAK2/STAT3 mediate
epithelial-mesenchymal transition in ovarian carcinomas. Br J
Cancer. 100:134–144. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Mayr D, Hirschmann A, Löhrs U and Diebold
J: KRAS and BRAF mutations in ovarian tumors: A comprehensive study
of invasive carcinomas, borderline tumors and extraovarian
implants. Gynecol Oncol. 103:883–887. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Hennessy B, Timms KM, Carey MS, Gutin A,
Meyer LA, Flake DD II, Abkevich V, Potter J, Pruss D, Glenn P, et
al: Somatic mutations in BRCA1 and BRCA2 could expand the number of
patients that benefit from poly (ADP ribose) polymerase inhibitors
in ovarian cancer. J Clin Oncol. 28:3570–3576. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Peng DX, Luo M, Qiu LW, He YL and Wang XF:
Prognostic implications of microRNA-100 and its functional roles in
human epithelial ovarian cancer. Oncol Rep. 27:1238–1244.
2012.PubMed/NCBI
|
|
8
|
Nakano H, Yamada Y, Miyazawa T and Yoshida
T: Gain-of-function microRNA screens identify miR-193a regulating
proliferation and apoptosis in epithelial ovarian cancer cells. Int
J Oncol. 42:1875–1882. 2013.PubMed/NCBI
|
|
9
|
Auersperg N, Wong AS, Choi KC, Kang SK and
Leung PC: Ovarian surface epithelium: Biology, endocrinology, and
pathology. Endocr Rev. 22:255–288. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Bowen NJ, Walker LD, Matyunina LV, Logani
S, Totten KA, Benigno BB and McDonald JF: Gene expression profiling
supports the hypothesis that human ovarian surface epithelia are
multipotent and capable of serving as ovarian cancer initiating
cells. BMC Med Genomics. 2:712009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lee YH, Kim JH, Zhou H, Kim BW and Wong
DT: Salivary transcriptomic biomarkers for detection of ovarian
cancer: For serous papillary adenocarcinoma. J Mol Med (Berl).
90:427–434. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Johnson WE, Li C and Rabinovic A:
Adjusting batch effects in microarray expression data using
empirical Bayes methods. Biostatistics. 8:118–127. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Leek JT and Storey JD: A general framework
for multiple testing dependence. Proc Natl Acad Sci USA. 105:pp.
18718–18723. 2008; View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Bolstad BM, Irizarry RA, Astrand M and
Speed TP: A comparison of normalization methods for high density
oligonucleotide array data based on variance and bias.
Bioinformatics. 19:185–193. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Smyth GK: Linear models and empirical
bayes methods for assessing differential expression in microarray
experiments. Stat Appl Genet Mol Biol. 3:Article32004. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Benjamini Y and Hochberg Y: Controlling
the false discovery rate: A practical and powerful approach to
multiple testing. J Royal Statist Society Series B Methodol.
57:289–300. 1995.
|
|
17
|
Huang DW, Sherman BT, Tan Q, Collins JR,
Alvord WG, Roayaei J, Stephens R, Baseler MW, Lane HC and Lempicki
RA: The DAVID gene functional classification tool: A novel
biological module-centric algorithm to functionally analyze large
gene lists. Genome Biol. 8:R1832007. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kanehisa M and Goto S: KEGG: Kyoto
encyclopedia of genes and genomes. Nucleic Acids Res. 28:27–30.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
von Mering C, Huynen M, Jaeggi D, Schmidt
S, Bork P and Snel B: STRING: A database of predicted functional
associations between proteins. Nucleic Acids Res. 31:258–261. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kohl M, Wiese S and Warscheid B:
Cytoscape: Software for visualization and analysis of biological
networksData Mining in Proteomics. Springer; pp. 291–303. 2011
|
|
21
|
He X and Zhang J: Why do hubs tend to be
essential in protein networks? PLoS Genet. 2:e882006. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Bader GD and Hogue CW: An automated method
for finding molecular complexes in large protein interaction
networks. BMC Bioinformatics. 4:22003. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Merico D, Isserlin R, Stueker O, Emili A
and Bader GD: Enrichment map: A network-based method for gene-set
enrichment visualization and interpretation. PLoS One.
5:e139842010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Karolchik D, Hinrichs AS, Furey TS, Roskin
KM, Sugnet CW, Haussler D and Kent WJ: The UCSC table browser data
retrieval tool. Nucleic Acids Res. 32:D493–D496. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Papadopoulos GL, Reczko M, Simossis VA,
Sethupathy P and Hatzigeorgiou AG: The database of experimentally
supported targets: A functional update of TarBase. Nucleic Acids
Res. 37:D155–D158. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Garcia DM, Baek D, Shin C, Bell GW,
Grimson A and Bartel DP: Weak seed-pairing stability and high
target-site abundance decrease the proficiency of lsy-6 and other
microRNAs. Nat Struct Mol Biol. 18:1139–1146. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Xiao F, Zuo Z, Cai G, Kang S, Gao X and Li
T: miRecords: An integrated resource for microRNA-target
interactions. Nucleic Acids Res. 37:D105–D110. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Sayer JA, Otto EA, O'Toole JF, Nurnberg G,
Kennedy MA, Becker C, Hennies HC, Helou J, Attanasio M, Fausett BV,
et al: The centrosomal protein nephrocystin-6 is mutated in Joubert
syndrome and activates transcription factor ATF4. Nat Genet.
38:674–681. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
29
|
Yuen KW, Montpetit B and Hieter P: The
kinetochore and cancer: What's the connection? Curr Opin Cell Biol.
17:576–582. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Ghosh-Choudhury T, Loomans HA, Wan YW, Liu
Z, Hawkins SM and Anderson ML: Abstract 3113: Hyperactivation of
FOXM1 drives ovarian cancer growth and metastasis independent of
the G2-M cell cycle checkpoint. Cancer Res. 73 Suppl:S31132013.
View Article : Google Scholar
|
|
31
|
Mercier PL, Bachvarova M, Plante M,
Gregoire J, Renaud MC, Ghani K, Têtu B, Bairati I and Bachvarov D:
Characterization of DOK1, a candidate tumor suppressor gene, in
epithelial ovarian cancer. Mol Oncol. 5:438–453. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Lee YC, Huang CC, Lin DY, Chang WC and Lee
KH: Overexpression of centromere protein K (CENPK) in ovarian
cancer is correlated with poor patient survival and associated with
predictive and prognostic relevance. PeerJ. 3:e13862015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Unoki M, Brunet J and Mousli M: Drug
discovery targeting epigenetic codes: The great potential of UHRF1,
which links DNA methylation and histone modifications, as a drug
target in cancers and toxoplasmosis. Biochem Pharmacol.
78:1279–1288. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Jenkins Y, Markovtsov V, Lang W, Sharma P,
Pearsall D, Warner J, Franci C, Huang B, Huang J, Yam GC, et al:
Critical role of the ubiquitin ligase activity of UHRF1, a nuclear
RING finger protein, in tumor cell growth. Mol Biol Cell.
16:5621–5629. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Yan F, Wang X, Shao L, Ge M and Hu X:
Analysis of UHRF1 expression in human ovarian cancer tissues and
its regulation in cancer cell growth. Tumor Biol. 36:8887–8893.
2015. View Article : Google Scholar
|
|
36
|
Mantovani R: The molecular biology of the
CCAAT-binding factor NF-Y. Gene. 239:15–27. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Gurtner A, Fuschi P, Magi F, Colussi C,
Gaetano C, Dobbelstein M, Sacchi A and Piaggio G: NF-Y dependent
epigenetic modifications discriminate between proliferating and
postmitotic tissue. PLoS One. 3:e20472008. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Yamanaka K, Mizuarai S, Eguchi T, Itadani
H, Hirai H and Kotani H: Expression levels of NF-Y target genes
changed by CDKN1B correlate with clinical prognosis in multiple
cancers. Genomics. 94:219–227. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Garipov A, Li H, Bitler BG, Thapa RJ,
Balachandran S and Zhang R: NF-YA underlies EZH2 upregulation and
is essential for proliferation of human epithelial ovarian cancer
cells. Mol Cancer Res. 11:360–369. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Zhang Y, Zhao FJ, Chen LL, Wang LQ, Nephew
KP, Wu YL and Zhang S: MiR-373 targeting of the Rab22a oncogene
suppresses tumor invasion and metastasis in ovarian cancer.
Oncotarget. 5:12291–12303. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Shen J, Ambrosone CB, DiCioccio RA, Odunsi
K, Lele SB and Zhao H: A functional polymorphism in the miR-146a
gene and age of familial breast/ovarian cancer diagnosis.
Carcinogenesis. 29:1963–1966. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Mullany LK, Liu Z, King ER, Wong KK and
Richards JS: Wild-type tumor repressor protein 53 (Trp53) promotes
ovarian cancer cell survival. Endocrinology. 153:1638–1648. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Wong KK, Lu KH, Malpica A, Bodurka DC,
Shvartsman HS, Schmandt RE, Thornton AD, Deavers MT, Silva EG and
Gershenson DM: Significantly greater expression of ER PR, and ECAD
in advanced-stage low-grade ovarian serous carcinoma as revealed by
immunohistochemical analysis. Int J Gynecol Pathol. 26:404–409.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Wong KK, Izaguirre DI, Kwan SY, King ER,
Deavers MT, Sood AK, Mok SC and Gershenson DM: Poor survival with
wild-type TP53 ovarian cancer? Gynecol Oncol. 130:565–569. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Storms RW, Trujillo AP, Springer JB, Shah
L, Colvin OM, Ludeman SM and Smith C: Isolation of primitive human
hematopoietic progenitors on the basis of aldehyde dehydrogenase
activity. Proc Natl Acad Sci USA. 96:pp. 9118–9123. 1999;
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Sun P, Sehouli J, Denkert C, Mustea A,
Könsgen D, Koch I, Wei L and Lichtenegger W: Expression of estrogen
receptor-related receptors, a subfamily of orphan nuclear
receptors, as new tumor biomarkers in ovarian cancer cells. J Mol
Med (Berl). 83:457–467. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Halon A, Materna V, Drag-Zalesinska M,
Nowak-Markwitz E, Gansukh T, Donizy P, Spaczynski M, Zabel M,
Dietel M, Lage H and Surowiak P: Estrogen receptor alpha expression
in ovarian cancer predicts longer overall survival. Pathol Oncol
Res. 17:511–518. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Liu Q, Chen Y, Pan JJ and Murakami T:
Expression of protocadherin-9 and protocadherin-17 in the nervous
system of the embryonic zebrafish. Gene Expr Patterns. 9:490–496.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Brasch J, Harrison OJ, Honig B and Shapiro
L: Thinking outside the cell: How cadherins drive adhesion. Trends
Cell Biol. 22:299–310. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Bendas G and Borsig L: Cancer cell
adhesion and metastasis: Selectins, integrins, and the inhibitory
potential of heparins. Int J Cell Biol. 2012:6767312012. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Asad M, Wong MK, Tan TZ, Choolani M, Low
J, Mori S, Virshup D, Thiery JP and Huang RY: FZD7 drives in vitro
aggressiveness in Stem-A subtype of ovarian cancer via regulation
of non-canonical Wnt/PCP pathway. Cell Death Dis. 5:e13462014.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Taylor DD and Gercel-Taylor C: MicroRNA
signatures of tumor-derived exosomes as diagnostic biomarkers of
ovarian cancer. Gynecol Oncol. 110:13–21. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Li Y, Yao L, Liu F, Hong J, Chen L, Zhang
B and Zhang W: Characterization of microRNA expression in serous
ovarian carcinoma. Int J Mol Med. 34:491–498. 2014.PubMed/NCBI
|