Introduction
Lung cancer is the leading cause of
cancer-associated mortality (accounting for ~13% of all diagnosed
cancers), with ~1.8 million new cases in 2012 (1). Between 2011 and 2013, the lung cancer
mortality rate increased by 464.84% in China, and this continued to
increase (2). Non-small cell lung
cancer (NSCLC) constitutes almost 85% of all lung cancers and is
usually diagnosed an advanced stage (3). Surgery followed by chemoradiotherapy is
the most commonly adopted method for locally advanced NSCLC, but
the efficacy is diverse across different patients (4). In previous years, immunotherapy became
an optional therapeutic for numerous types of cancers, including
melanoma and lung cancer, through sustained activation of the
immune system (5,6). Certain immune checkpoint inhibitors have
been shown to significantly improve the prognosis of a number of
cancers, including cytotoxic T-lymphocyte antigen 4, and nivolumab
may prolong the median survival time of patients with melanoma and
NSCLC by 2–4 and 6–9 months, respectively (7). However, the morbidity and mortality rate
of NSCLC remains high, and additional molecular biology studies are
required for more precise treatment and earlier diagnosis.
Next generation RNA sequencing (RNA-Seq) is becoming
a popular technology for the quantification of RNA, including mRNA
(such as microRNA) and noncoding RNA (such as long noncoding RNA).
Variation of genomic structure, including alternative splicing (AS)
and single nucleotide polymorphisms, can also be detected by
RNA-Seq. Compared with gene microarray, the most evident advantage
of RNA-Seq is its ability to detect the expression values of low
abundance as well as novel transcripts (8). Previous studies of NSCLC, based on
RNA-Seq have been conducted and successfully identified numerous
potential targets, involved in the progression or suppression of
cancer. Riccardo et al identified the adenosine A3 receptor
as a valuable target for NSCLC, based on the combination of gene
fusion and differential expression analysis through RNA-Seq
(9). In the previous study by Han
et al, via RNA-Seq analysis, chromobox 3 and cellular
retinoic acid-binding protein 2 were also considered to contribute
to the progression of NSCLC (10).
Compared with differential expression analysis at
the gene level, exon level analysis may improve analysis of disease
stages (11). The present study aimed
to identify the potential targets of NSCLC, through the combined
analysis of differential expression and AS, and based on the
RNA-Seq dataset from the Gene Expression Omnibus (GEO). This may be
helpful for the precise treatment of NSCLC, as well as early
diagnosis.
Materials and methods
RNA-Seq dataset
The RNA-Seq dataset (GSE68795) in the present study
was obtained from GEO (http://www.ncbi.nlm.nih.gov/geo/), which was
accumulated by Durrans et al (12). A total of 17 samples were included,
containing three immature monocytic myeloid cell (IMMC) samples,
two epithelial cell (Epi) samples and two neutrophil (Neu) samples
from lung cancer patients, as well as three IMMC samples, three Epi
samples and four Neu samples from adjacent normal lung tissues.
Illumina HiSeq 2000 (Illumina, Inc., San Diego, CA, USA) was used
for the sequencing process. Briefly, total RNA was extracted from
flow cytometry sorted cells; TruSeq RNA Sample Preparation kit
(Illumina, Inc.) was used for the preparation of cDNA libraries
from 15–35 ng RNA; cDNA libraries that passed size and purity check
were retained for the following sequencing. Single-end 51 bp short
sequences (reads) were generated for the IMMC and Neu samples,
while paired-end 102 bp reads were generated for the Epi samples,
in lung cancer and adjacent normal lung tissues.
Reads mapping and assembling
Quality control of raw reads was conducted using
FastQC software version 1.3, which was developed by Andrews
(13), with the default parameters,
i.e., reads with a quality score <10 and N >5% were
discarded. The remaining reads were mapped to the UCSC genome
(version GRCh37/hg19) through TopHat (2.1.0.Linux_x86_64) (14), a fast splice junction mapper for
RNA-Seq reads, with no more than 2 mismatches in 25 bp segments.
Cufflinks (14) (2.2.1.Linux_x86_64)
was used for the assembly of the mapped reads, which allowed for
the identification of novel transcripts, and fragments per kilobase
of exon per million fragments mapped (FPKM) representation of gene
expression value was obtained.
Differential expression analysis
Cuffdiff of Cufflinks was used to test the
significance of differential expression of genes based on FPKM.
Genes with fold change (FC) >2 (upregulated) or <0.5
(downregulated), and false discovery rate (FDR) adjusted for
P<0.05, were considered to be differentially expressed.
Differential alternative splicing
analysis
The replicate multivariate analysis of transcript
splicing (rMATS) (15), developed by
Shen et al, was used to screen differential alternative splicing
genes across samples. The mapping results (in bam format) were
submitted to rMATS. Annotation of genes (in GTF format) was
obtained from the UCSC and used for the screening of known splicing
sites. Finally, five main alternative splicing types, including
skipping exon (SE), retention intron (RI), alternative 5′splice
site (A5SS), alternative 3′splice site (A3SS) and mutually
exclusive exons (MXE), which satisfied the criteria of FDR <0.1,
were screened out as DAS genes.
Functional enrichment analysis
The DE and DAS genes were submitted to the Database
for Annotation, Visualization and Integrated Discovery (DAVID;
https://david.ncifcrf.gov/) (16) for the analysis of enrichment of gene
ontology (GO) terms and Kyoto Encyclopedia of Genes and Genomes
(KEGG) pathways. P<0.05 was considered to indicate a
statistically significant difference for the screening of
significant GO terms and pathways.
Results
RNA-Seq landscape
The average number of reads obtained from the
RNA-Seq dataset was 60242792, and at least 35 million reads were
prepared for every sample, as shown in Table I. The clean reads were mapped to the
human genome (hg19) by TopHat and 86.50–91.90% unique mapped rates
were obtained across all samples. These were used for the following
analysis.
 | Table I.Mapping overview of all the
RNA-Sequencing experiments. |
Table I.
Mapping overview of all the
RNA-Sequencing experiments.
| Sample | All reads | Mapped reads | Mapped rates, % | Unique mapped | Unique mapped rates,
% |
|---|
| Adj-Epi1 |
97717750 |
92115270 | 94.3 |
87061215 | 89.1 |
| Adj-Epi2 |
63907228 |
58725212 | 91.9 |
55272496 | 86.5 |
| Adj-Epi3 | 123019030 | 116594941 | 94.8 | 110088025 | 89.5 |
| Adj-IMM1 |
40304691 |
38809145 | 96.3 |
37030002 | 91.9 |
| Adj-IMM2 |
36685339 |
35109464 | 95.7 |
33284563 | 90.7 |
| Adj-IMM3 |
44679997 |
42067287 | 94.2 |
40025779 | 89.6 |
| Adj-Neu1 |
49967221 |
47183219 | 94.4 |
45452825 | 91.0 |
| Adj-Neu2 |
50458780 |
47159700 | 93.5 |
44872985 | 88.9 |
| Adj-Neu3 |
53402957 |
50511053 | 94.6 |
48090506 | 90.1 |
| Adj-Neu4 |
52277703 |
49603994 | 94.9 |
47551180 | 91.0 |
| Tum-Epi1 | 106711340 | 101588228 | 95.2 |
95969883 | 89.9 |
| Tum-Epi2 |
94931814 |
90391800 | 95.2 |
85487063 | 90.1 |
| Tum-IMM1 |
45344289 |
43038591 | 94.9 |
41037840 | 90.5 |
| Tum-IMM2 |
42060455 |
40620197 | 96.6 |
38638529 | 91.9 |
| Tum-IMM3 |
35570701 |
33745251 | 94.9 |
32229614 | 90.6 |
| Tum-Neu1 |
37106424 |
34076868 | 91.8 |
32442881 | 87.4 |
| Tum-Neu2 |
49981740 |
47119502 | 94.3 |
44980801 | 90.0 |
Differential expression analysis
The unique mapped reads were assembled into
transcripts by cufflinks, and FPKM values were obtained to quantify
the mRNA expression level. Based on the criteria of FC >2 or
<0.5, and FDR adjusted P<0.05, umerous genes were revealed to
be differential expression in tumor samples compared with normal
samples.
In Epi tumor samples, a total of 1,624 DE genes were
identified, which contained 915 downregulated and 709 upregulated
genes. However, in IMMC tumor samples, 1,593 DE genes (639
downregulated and 954 upregulated genes) were obtained. In Neu
tumor samples, 1,672 DE genes were obtained, which consisted of
1,141 downregulated and 1,672 upregulated genes. Furthermore, as
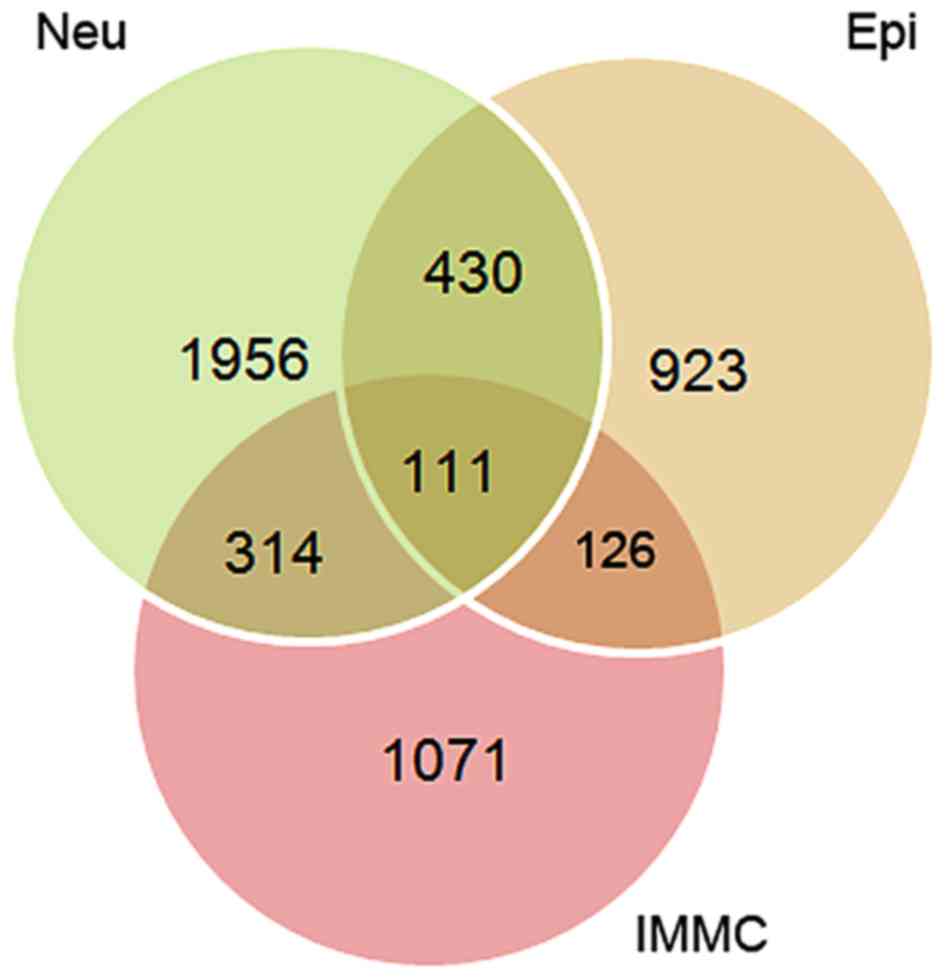
shown in Fig. 1, 111 overlapping
genes were identified among those three groups of DE genes, and 91
of those overlaps were consistently upregulated or downregulated
across all three types of NSCLC tumor cells.
AS in NSCLC tumor samples
Through AS, a mRNA precursor may result in multiple
mature mRNAs with different functions. In the present study, the
significant DAS genes were identified for the tumor samples
compared with the normal ones through rMATS, with the threshold of
FDR <0.1. Notably, alternative splicing of genes in Neu tumor
samples was not found to be significantly different compared with
Neu normal samples. A total of 2,130 and 1,713 DAS genes were
identified in Epi and IMMC tumor samples, respectively. In total,
272 overlapping genes were screened out between the two lists of
DAS genes.
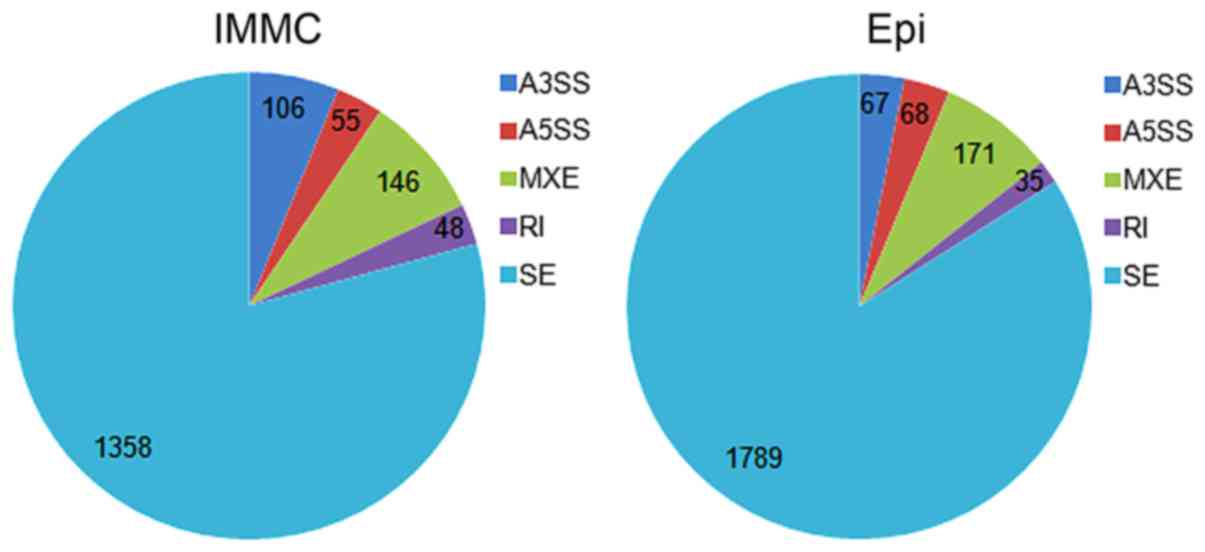
A total of 5 types of AS events were identified,
including A3SS, A5SS, MXE, RI and SE. As shown in Fig. 2, SE is the most prevalent AS event in
Epi and IMMC tumor samples, and accounts for 84.00 and 79.28% of
all the AS events in Epi and IMMC, respectively. RI is the least
prevalent AS event, which only accounts for 1.64 and 2.80% of all
the AS events in Epi and IMMC, respectively.
GO and pathway analysis
Functional enrichment analysis based on the DAVID
identified a number of significantly enriched GO terms and KEGG
pathways in DE and DAS genes. The top five most significant GO
terms of DE genes of the three groups of NSCLC tumor samples are
listed in Table II. As shown in
Table II, biological processes
associated with the cell and immune system were significantly
enriched (P<0.05) in the DE genes of all the three types of
NSCLC tumor samples. Enriched pathways of DE genes of Epi, IMMC and
Neu tumor samples are listed in Tables
III, IV and V, respectively. Similar to the enriched GO
terms, those pathways were mainly involved in the dysregulation of
cell division and abnormal activity of the immune and inflammatory
systems.
 | Table II.Top five most significantly enriched
GO terms of differential expression genes of the three groups of
non-small cell lung cancer samples. |
Table II.
Top five most significantly enriched
GO terms of differential expression genes of the three groups of
non-small cell lung cancer samples.
| Category | GO name | P-value |
|---|
| Epi samples |
|
|
| BP | Response to
wounding |
9.39×10−15 |
| CC | Extracellular
space |
8.69×10−14 |
| BP | Cell adhesion |
2.36×10−11 |
| BP | Biological
adhesion |
2.59×10−11 |
| CC | Cell fraction |
6.12×10−11 |
| IMMC samples |
|
|
| BP | Immune
response |
3.22×10−10 |
| BP | Response to
wounding |
3.36×10−7 |
| BP | Cell division |
2.27×10−6 |
| CC | Plasma membrane
part |
6.03×10−6 |
| BP | Mitosis |
6.98×10−6 |
| Neu samples |
|
|
| BP | Cell adhesion |
1.29×10−10 |
| BP | Biological
adhesion |
1.50×10−10 |
| MF | GTPase regulator
activity |
1.90×10−10 |
| MF |
Nucleoside-triphosphatase |
4.24×10−10 |
| CC | Plasma membrane
part |
5.47×10−10 |
 | Table III.Kyoto Encyclopedia of Genes and
Genomes pathways of differential expression genes in epithelial
cell tumor samples. |
Table III.
Kyoto Encyclopedia of Genes and
Genomes pathways of differential expression genes in epithelial
cell tumor samples.
| Pathway name | Gene number | P-value |
|---|
| ECM-receptor
interaction | 29 |
1.64×10−9 |
| Complement and
coagulation cascades | 21 |
4.45×10−6 |
| Biosynthesis of
unsaturated fatty acids | 10 |
1.09×10−4 |
| Dilated
cardiomyopathy | 20 |
1.04×10−3 |
| Focal adhesion | 34 |
1.51×10−3 |
| PPAR signaling
pathway | 16 |
2.02×10−3 |
| Hematopoietic cell
lineage | 18 |
3.08×10−3 |
| Pathways in
cancer | 48 |
3.29×10−3 |
| Prion diseases | 10 |
4.92×10−3 |
| Hypertrophic
cardiomyopathy | 17 |
6.63×10−3 |
| Arrhythmogenic
right ventricular cardiomyopathy | 15 |
1.30×10−2 |
| Cell adhesion
molecules | 22 |
1.47×10−2 |
| Ether lipid
metabolism | 9 |
1.63×10−2 |
| Systemic lupus
erythematosus | 17 |
2.70×10−2 |
| Basal cell
carcinoma | 11 |
3.58×10−2 |
| MAPK signaling
pathway | 36 |
3.66×10−2 |
| Synthesis and
degradation of ketone bodies | 4 |
4.86×10−2 |
| Terpenoid backbone
biosynthesis | 5 |
4.99×10−2 |
 | Table IV.Kyoto Encyclopedia of Genes and
Genomes pathways of differential expression genes in immature
monocytic myeloid cell tumor samples. |
Table IV.
Kyoto Encyclopedia of Genes and
Genomes pathways of differential expression genes in immature
monocytic myeloid cell tumor samples.
| Pathway name | Gene number | P-value |
|---|
| Hematopoietic cell
lineage | 25 |
7.50×10−7 |
| DNA
replication | 13 |
7.02×10−5 |
| Cell cycle | 26 |
2.25×10−4 |
| Cytokine-cytokine
receptor interaction | 42 |
7.78×10−4 |
| Cell adhesion
molecules | 25 |
1.26×10−3 |
| ECM-receptor
interaction | 18 |
1.95×10−3 |
| Purine
metabolism | 25 |
9.05×10−3 |
| Chemokine signaling
pathway | 29 |
9.52×10−3 |
| Pantothenate and
CoA biosynthesis | 6 |
1.02×10−2 |
| Aminoacyl-tRNA
biosynthesis | 10 |
1.29×10−2 |
| Bladder cancer | 10 |
1.51×10−2 |
| PPAR signaling
pathway | 13 |
2.75×10−2 |
| Pyrimidine
metabolism | 16 |
3.30×10−2 |
| Viral
myocarditis | 13 |
3.36×10−2 |
| Axon guidance | 20 |
3.44×10−2 |
| Pathways in
cancer | 42 |
3.80×10−2 |
| Focal adhesion | 28 |
3.92×10−2 |
 | Table V.Kyoto Encyclopedia of Genes and
Genomes pathways of differential expression genes in neutrophil
tumor samples. |
Table V.
Kyoto Encyclopedia of Genes and
Genomes pathways of differential expression genes in neutrophil
tumor samples.
| Pathway name | Gene number | P-value |
|---|
| Focal adhesion | 61 |
4.81×10−7 |
| Regulation of actin
cytoskeleton | 62 |
2.59×10−6 |
| Fc gamma R-mediated
phagocytosis | 34 |
5.94×10−6 |
| Chemokine signaling
pathway | 53 |
2.67×10−5 |
| Leukocyte
transendothelial migration | 37 |
5.92×10−5 |
| ECM-receptor
interaction | 29 |
7.03×10−5 |
| Axon guidance | 37 |
4.28×10−4 |
| Pathogenic
Escherichia coli infection | 20 |
1.01×10−3 |
| Hypertrophic
cardiomyopathy | 26 |
1.40×10−3 |
| Adherens
junction | 24 |
1.71×10−3 |
| Pathways in
cancer | 73 |
2.62×10−3 |
| Dilated
cardiomyopathy | 26 |
4.57×10−3 |
| Valine, leucine and
isoleucine degradation | 15 |
7.22×10−3 |
| Glioma | 19 |
8.70×10−3 |
| Natural killer cell
mediated cytotoxicity | 32 | 0.018518 |
| Non-small cell lung
cancer | 16 |
2.06×10−2 |
| Gap junction | 23 |
2.30×10−2 |
|
Aldosterone-regulated sodium
reabsorption | 13 |
2.45×10−2 |
| Endocytosis | 41 |
2.55×10−2 |
| MAPK signaling
pathway | 56 |
2.78×10−2 |
|
Progesterone-mediated oocyte
maturation | 22 |
2.95×10−2 |
| Arrhythmogenic
right ventricular cardiomyopathy | 20 |
2.97×10−2 |
| ErbB signaling
pathway | 22 |
3.32×10−2 |
| Fc epsilon RI
signaling pathway | 20 |
3.81×10−2 |
| p53 signaling
pathway | 18 |
3.84×10−2 |
| Colorectal
cancer | 21 |
4.23×10−2 |
| Cell adhesion
molecules | 30 |
4.58×10−2 |
| Glycerolipid
metabolism | 13 |
4.84×10−2 |
| B cell receptor
signaling pathway | 19 |
4.89×10−2 |
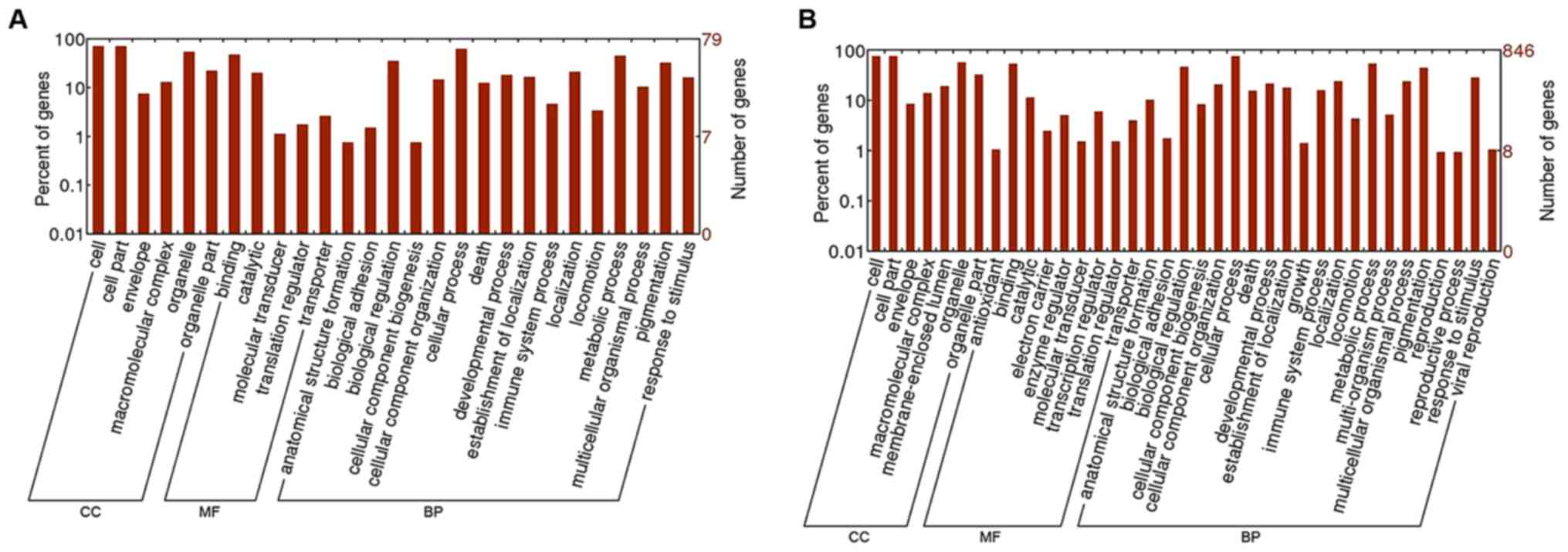
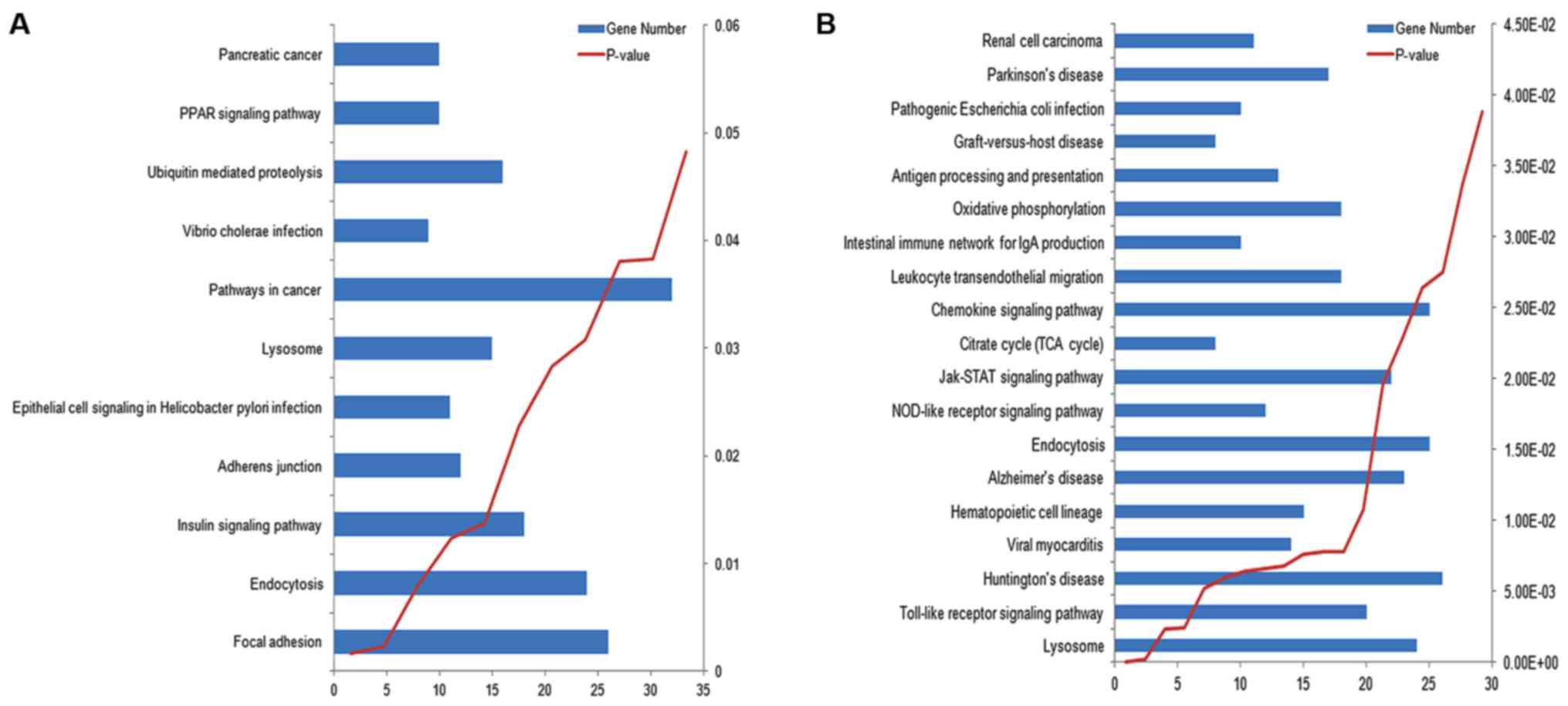
For DAS genes in Epi and IMMC tumor samples, their
enriched GO terms are shown in Fig.
3, which was obtained by submitting the DAVID result to the
WEGO website (http://wego.genomics.org.cn/cgi-bin/wego/index.pl),
and their enriched KEGG pathways are shown in Fig. 4. As expected, those DAS genes were
also involved in cell and immune abnormality. The insulin signaling
pathway and oxidative phosphorylation, which were associated with
substance metabolism and were not revealed by DE genes, were also
found to be significantly enriched (P<0.05) in Epi and IMMC
tumor samples, respectively. DE and DAS genes may be complementary
in the understanding of mechanisms of NSCLC.
Discussion
Lung cancer remains the leading cause of
cancer-associated mortality in recent years, and its main type is
NSCLC (17). An increasing number of
studies about NSCLC have been emerging and numerous potential
biomarkers for the early diagnosis or treatment of NSCLC have been
identified subsequent to the development of molecular biology
technologies. However, the mechanisms of NSCLC remain largely
unknown, and additional studies are required to improve its
prognosis. In the present study, through re-analysis of the RNA-Seq
dataset from the GEO database, the significant DE and DAS genes
were screened out in three types of NSCLC tumor samples (Epi, IMMC
and Neu), compared with the corresponding normal samples, and their
enriched GO terms and KEGG pathways were obtained. The present
study aimed to provide results to help with the development of
therapeutic methods and drugs for NSCLC.
The rapid growth of RNA-Seq provides the chance to
observe variation in the genome at the exon level, and screening of
alternative splicing is one of its important applications (18). AS is an important mechanism that could
result in proteins with different functions from one mRNA
precursor, and it has been shown to affect the status of numerous
diseases. AS events involving exons 15–19 of d'Origine
nantais tyrosine kinase have been screened out in lung cancer
(19). In addition, the T cell factor
4K AS isoform was found to improve the prognosis of NSCLC by
suppressing the proliferation and metastasis of tumor cells
(20). In the present study,
significant alternative splicing was not observed in any genes in
the Neu tumor samples compared with the normal samples. Numerous AS
genes were identified in Epi and IMMC tumor samples, which may
indicate the differences of those three types of cells in the
inducement of NSCLC.
In accordance with previous studies, SE is the most
prevalent AS event, while RI is the least prevalent AS event in Epi
and IMMC tumor samples (21,22). A total of 272 genes demonstrated
alternative splicing in Epi and IMMC tumor samples, and this may be
an indication of the important roles of those genes in the
progression of NSCLC. Compared with AS analysis, numerous DE genes
were identified in all three types of tumor samples, and 111
overlapping genes were screened out. One of the significant
differences between the DE genes and DAS genes is the involvement
of substance metabolism-associated pathways of DAS genes, including
the insulin signaling pathway and oxidative phosphorylation.
Insulin-like growth factor (IGF-1R) performs important roles
through its downstream signaling in a number of solid tumors,
including NSCLC (23). Furthermore,
IGF-1R affects cisplatin and radiation resistance in lung cancer
(24). In addition, oxidative
stress/activity is affected by IGF-1R in numerous diseases,
resulting in the dysregulation of biosystems, such as in cancer or
neurodegeneration (25,26). DE and DAS analysis revealed a number
of common GO terms and pathways, and the primary terms and pathways
were cell or immune-associated. These have been investigated in
numerous studies and were confirmed to play important roles in the
development of numerous types of cancers, including lung cancer,
gastric cancer and liver cancer (27–29).
Based on the integrated analysis of DE and DAS,
alternative splicing was identified by leucine-rich repeat kinase 2
(LRRK2) in Epi and IMMC tumor samples, and was consistently
downregulated in all three types of tumor cells. LRRK2 is a member
of the leucine-rich repeat kinase family and encodes a protein with
an ankryin repeat region. LRRK2 has been shown to play important
roles in neurodegeneration diseases, including Parkinson's disease
(30–32). The role of LRRK2 in cancer development
remains unclear; however, certain indirect associations were
identified (33). For example,
through disrupting the activation of the proto-oncogene MET, LRRK2
may prevent tumor cell proliferation (34). Therefore, it may be a novel target for
NSCLC.
In conclusion, through the combined analysis of DE
and DAS, potential targets for NSCLC were screened out.
Furthermore, DE and DAS may be complementary for understanding the
mechanisms of NSCLC. The present study may be helpful in the
diagnosis and treatment of NSCLC.
References
|
1
|
He X, Wang J and Li Y: Efficacy and safety
of docetaxel for advanced non-small-cell lung cancer: A
meta-analysis of Phase III randomized controlled trials. Onco
Targets Ther. 8:2023–2031. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
She J, Yang P, Hong Q and Bai C: Lung
cancer in China: Challenges and interventions. Chest.
143:1117–1126. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Santarpia M, González-Cao M, Viteri S,
Karachaliou N, Altavilla G and Rosell R: Programmed cell death
protein-1/programmed cell death ligand-1 pathway inhibition and
predictive biomarkers: Understanding transforming growth
factor-beta role. Transl Lung Cancer Res. 4:728–742.
2015.PubMed/NCBI
|
|
4
|
Nakajima Y, Akiyama H, Kinoshita H, Atari
M, Fukuhara M, Saito Y, Sakai H and Uramoto H: Case report of two
patients having successful surgery for lung cancer after treatment
for Grade 2 radiation pneumonitis. Ann Med Surg (Lond). 5:1–4.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wieder T, Brenner E, Braumüller H and
Röcken M: Immunotherapy of melanoma: Efficacy and mode of action. J
Dtsch Dermatol Ges. 14:28–37. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Pilotto S, Molina-Vila MA, Karachaliou N,
Carbognin L, Viteri S, González-Cao M, Bria E, Tortora G and Rosell
R: Integrating the molecular background of targeted therapy and
immunotherapy in lung cancer: A way to explore the impact of
mutational landscape on tumor immunogenicity. Transl Lung Cancer
Res. 4:721–727. 2015.PubMed/NCBI
|
|
7
|
Kobold S, Duewell P, Schnurr M, Subklewe
M, Rothenfusser S and Endres S: Immunotherapy in tumors. Dtsch
Arztebl Int. 112:809–815. 2015.PubMed/NCBI
|
|
8
|
Morozova O and Marra MA: Applications of
next-generation sequencing technologies in functional genomics.
Genomics. 92:255–264. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Riccardo F, Arigoni M, Buson G, Zago E,
Iezzi M, Longo D, Carrara M, Fiore A, Nuzzo S, Bicciato S, et al:
Characterization of a genetic mouse model of lung cancer: A promise
to identify non-small cell lung cancer therapeutic targets and
biomarkers. BMC Genomics. 15 Suppl 3:12014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Han SS, Kim WJ, Hong Y, Hong SH, Lee SJ,
Ryu DR, Lee W, Cho YH, Lee S, Ryu YJ, et al: RNA sequencing
identifies novel markers of non-small cell lung cancer. Lung
Cancer. 84:229–235. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Bisognin A, Pizzini S, Perilli L, Esposito
G, Mocellin S, Nitti D, Zanovello P, Bortoluzzi S and Mandruzzato
S: An integrative framework identifies alternative splicing events
in colorectal cancer development. Mol Oncol. 8:129–141. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Durrans A, Gao D, Gupta R, Fischer KR,
Choi H, El Rayes T, Ryu S, Nasar A, Spinelli CF, Andrews W, et al:
Identification of reprogrammed myeloid cell transcriptomes in
NSCLC. PLoS One. 10:e01291232015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Andrews S: FastQC: A quality control tool
for high throughput sequence data. http://www.bioinformatics.babraham.ac.uk/projects/fastqc/January
10–2016
|
|
14
|
Trapnell C, Roberts A, Goff L, Pertea G,
Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL and Pachter L:
Differential gene and transcript expression analysis of RNA-seq
experiments with TopHat and Cufflinks. Nat Protoc. 7:562–578. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Shen S, Park JW, Lu ZX, Lin L, Henry MD,
Wu YN, Zhou Q and Xing Y: rMATS: Robust and flexible detection of
differential alternative splicing from replicate RNA-Seq data. Proc
Natl Acad Sci USA. 111:pp. E5593–E5601. 2014; View Article : Google Scholar : PubMed/NCBI
|
|
16
|
da W Huang, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009.PubMed/NCBI
|
|
17
|
Sang Y, Bi X, Liu Y, Zhang W and Wang D:
Adverse prognostic impact of TGFB1 T869C polymorphism in
non-small-cell lung cancer. Onco Targets Ther. 10:1513–1518. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Han Y, Gao S, Muegge K, Zhang W and Zhou
B: Advanced applications of RNA sequencing and challenges.
Bioinform Biol Insights. 9 Suppl 1:29–46. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Krishnaswamy S, Mohammed AK, Amer OE,
Tripathi G, Alokail MS and Al-Daghri NM: Novel splicing variants of
recepteur d'origine nantais (RON) tyrosine kinase involving exons
15–19 in lung cancer. Lung Cancer. 92:41–46. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Fan YC, Min L, Chen H and Liu YL:
Alternative splicing isoform of T cell factor 4K suppresses the
proliferation and metastasis of non-small cell lung cancer cells.
Genet Mol Res. 14:14009–14018. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wang X, Xu X, Lu X, Zhang Y and Pan W:
Transcriptome Bioinformatical analysis of vertebrate stages of
Schistosoma japonicum reveals alternative splicing events. PLoS
One. 10:e01384702015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Hide WA, Babenko VN, van Heusden PA,
Seoighe C and Kelso JF: The contribution of exon-skipping events on
chromosome 22 to protein coding diversity. Genome Res.
11:1848–1853. 2001.PubMed/NCBI
|
|
23
|
Scagliotti GV and Novello S: The role of
the insulin-like growth factor signaling pathway in non-small cell
lung cancer and other solid tumors. Cancer Treat Rev. 38:292–302.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Sun Y, Zheng S, Torossian A, Speirs CK,
Schleicher S, Giacalone NJ, Carbone DP, Zhao Z and Lu B: Role of
insulin-like growth factor-1 signaling pathway in
cisplatin-resistant lung cancer cells. Int J Radiat Oncol Biol
Phys. 82:e563–e572. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Dávila D, Fernández S and Torres-Alemán I:
Astrocyte resilience to oxidative stress induced by insulin-like
growth factor I (IGF-I) involves preserved AKT (protein kinase B)
activity. J Biol Chem. 291:2510–2523. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Khawaja T, Chokshi A, Ji R, Kato TS, Xu K,
Zizola C, Wu C, Forman DE, Ota T, Kennel P, et al: Ventricular
assist device implantation improves skeletal muscle function,
oxidative capacity, and growth hormone/insulin-like growth factor-1
axis signaling in patients with advanced heart failure. J Cachexia
Sarcopenia Muscle. 5:297–305. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
De Palma M: The role of the immune system
in cancer: From mechanisms to clinical applications. Biochim
Biophys Acta. 1865:1–2. 2016.PubMed/NCBI
|
|
28
|
Soto-Ortiz L and Finley SD: A cancer
treatment based on synergy between anti-angiogenic and immune cell
therapies. J Theor Biol. 394:197–211. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Boros LG, D'Agostino DP, Katz HE, Roth JP,
Meuillet EJ and Somlyai G: Submolecular regulation of cell
transformation by deuterium depleting water exchange reactions in
the tricarboxylic acid substrate cycle. Med Hypotheses. 87:69–74.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Oosterveld LP, Allen JC Jr, Ng EY, Seah
SH, Tay KY, Au WL, Tan EK and Tan LC: Greater motor progression in
patients with Parkinson disease who carry LRRK2 risk variants.
Neurology. 85:1039–1042. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Marder K, Wang Y, Alcalay RN,
Mejia-Santana H, Tang MX, Lee A, Raymond D, Mirelman A,
Saunders-Pullman R, Clark L, et al: Age-specific penetrance of
LRRK2 G2019S in the Michael J. Fox Ashkenazi Jewish LRRK2
consortium. Neurology. 85:89–95. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Lee JW and Cannon JR: LRRK2 mutations and
neurotoxicant susceptibility. Exp Biol Med (Maywood). 240:752–759.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Bae JR and Lee BD: Function and
dysfunction of leucine-rich repeat kinase 2 (LRRK2): Parkinson's
disease and beyond. BMB Reports. 48:243–248. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Looyenga BD, Furge KA, Dykema KJ, Koeman
J, Swiatek PJ, Giordano TJ, West AB, Resau JH, Teh BT and MacKeigan
JP: Chromosomal amplification of leucine-rich repeat kinase-2
(LRRK2) is required for oncogenic MET signaling in papillary renal
and thyroid carcinomas. Proc Natl Acad Sci USA. 108:pp. 1439–1444.
2011; View Article : Google Scholar : PubMed/NCBI
|