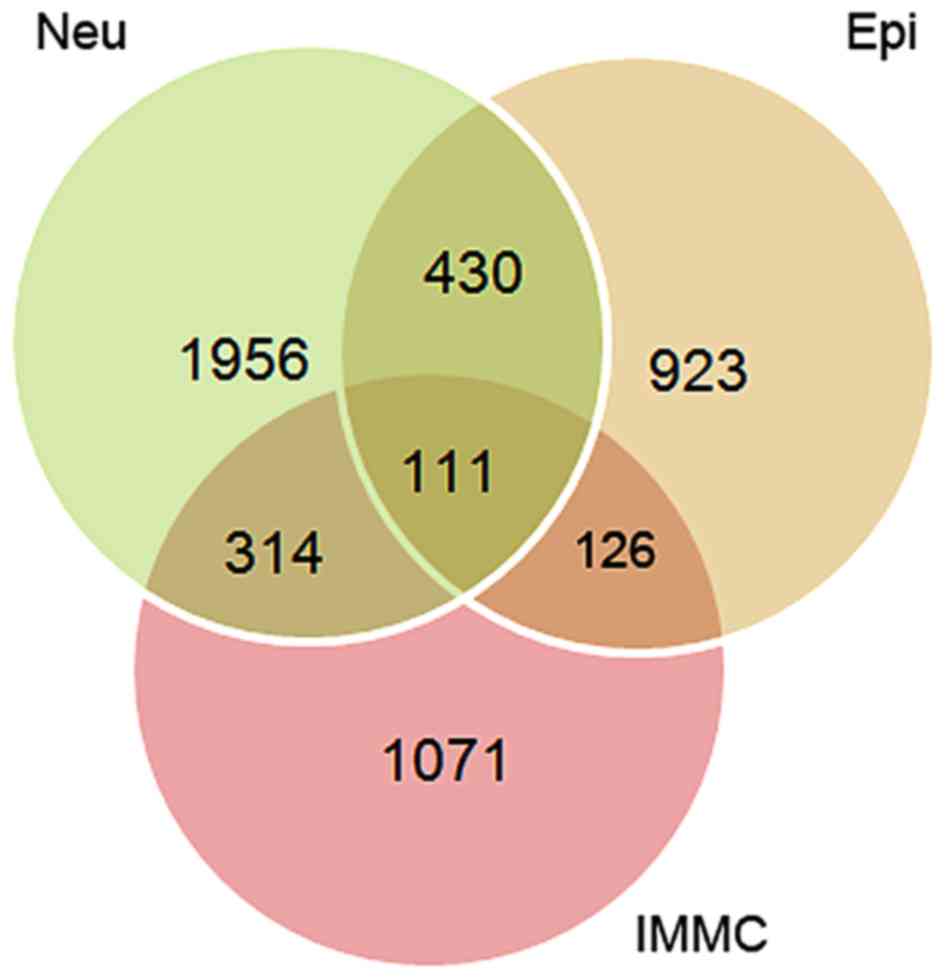
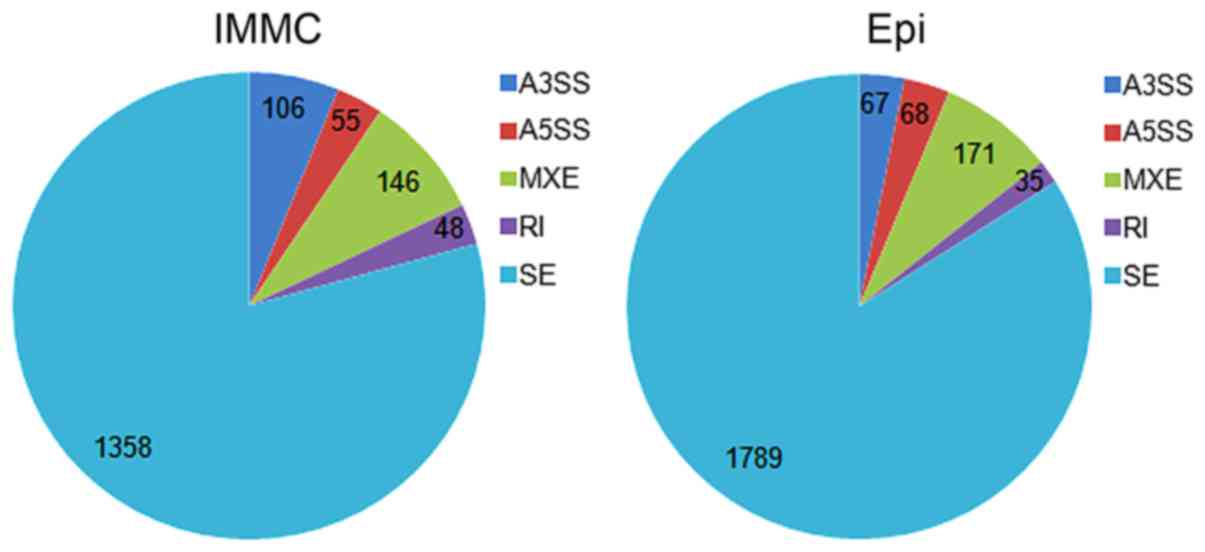
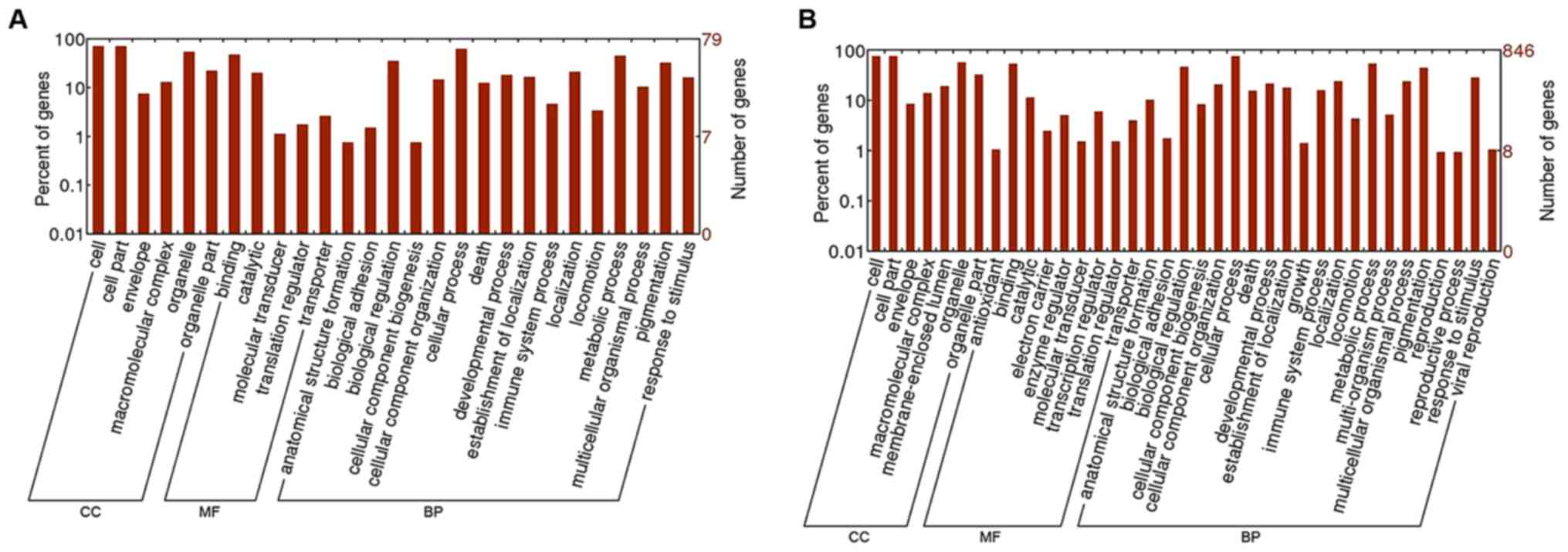
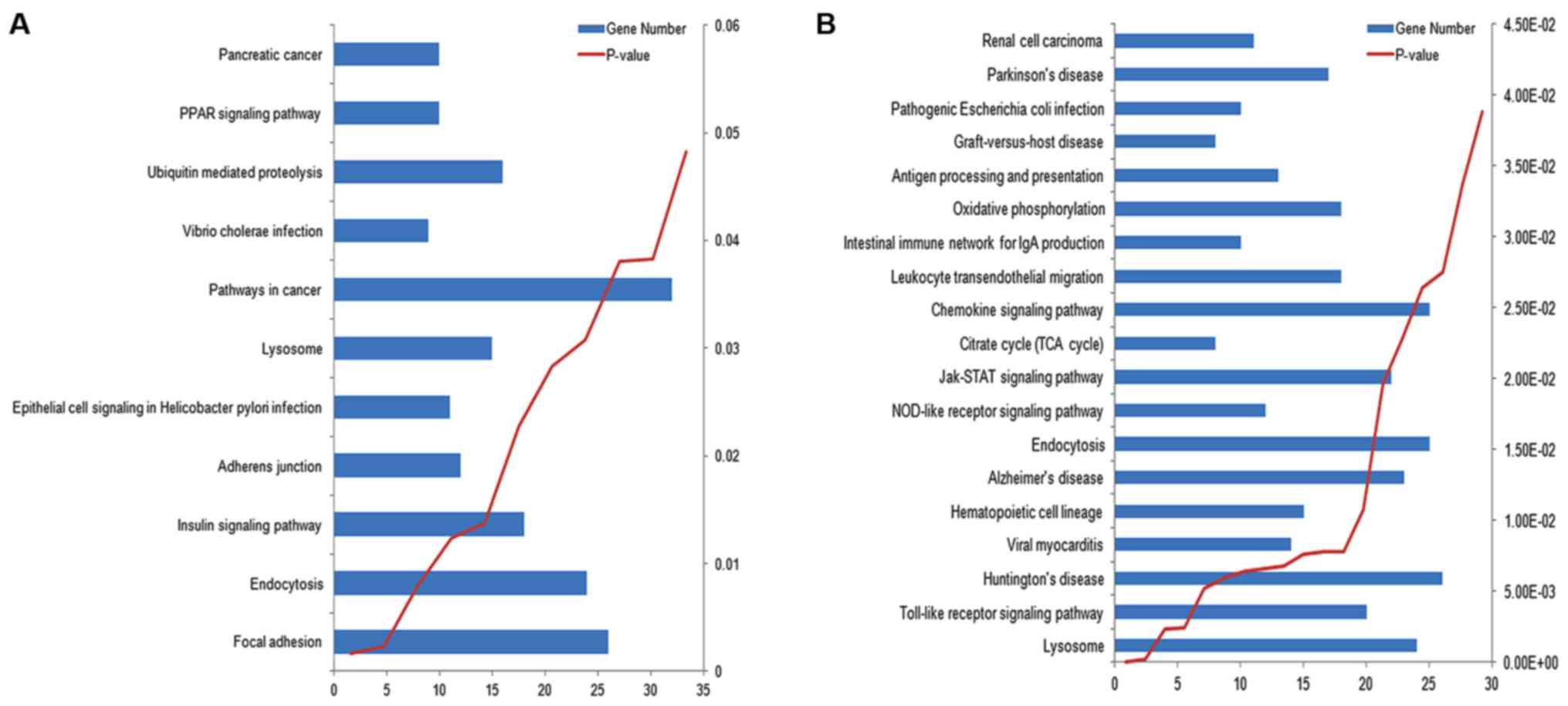
|
1
|
He X, Wang J and Li Y: Efficacy and safety
of docetaxel for advanced non-small-cell lung cancer: A
meta-analysis of Phase III randomized controlled trials. Onco
Targets Ther. 8:2023–2031. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
She J, Yang P, Hong Q and Bai C: Lung
cancer in China: Challenges and interventions. Chest.
143:1117–1126. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Santarpia M, González-Cao M, Viteri S,
Karachaliou N, Altavilla G and Rosell R: Programmed cell death
protein-1/programmed cell death ligand-1 pathway inhibition and
predictive biomarkers: Understanding transforming growth
factor-beta role. Transl Lung Cancer Res. 4:728–742.
2015.PubMed/NCBI
|
|
4
|
Nakajima Y, Akiyama H, Kinoshita H, Atari
M, Fukuhara M, Saito Y, Sakai H and Uramoto H: Case report of two
patients having successful surgery for lung cancer after treatment
for Grade 2 radiation pneumonitis. Ann Med Surg (Lond). 5:1–4.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wieder T, Brenner E, Braumüller H and
Röcken M: Immunotherapy of melanoma: Efficacy and mode of action. J
Dtsch Dermatol Ges. 14:28–37. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Pilotto S, Molina-Vila MA, Karachaliou N,
Carbognin L, Viteri S, González-Cao M, Bria E, Tortora G and Rosell
R: Integrating the molecular background of targeted therapy and
immunotherapy in lung cancer: A way to explore the impact of
mutational landscape on tumor immunogenicity. Transl Lung Cancer
Res. 4:721–727. 2015.PubMed/NCBI
|
|
7
|
Kobold S, Duewell P, Schnurr M, Subklewe
M, Rothenfusser S and Endres S: Immunotherapy in tumors. Dtsch
Arztebl Int. 112:809–815. 2015.PubMed/NCBI
|
|
8
|
Morozova O and Marra MA: Applications of
next-generation sequencing technologies in functional genomics.
Genomics. 92:255–264. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Riccardo F, Arigoni M, Buson G, Zago E,
Iezzi M, Longo D, Carrara M, Fiore A, Nuzzo S, Bicciato S, et al:
Characterization of a genetic mouse model of lung cancer: A promise
to identify non-small cell lung cancer therapeutic targets and
biomarkers. BMC Genomics. 15 Suppl 3:12014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Han SS, Kim WJ, Hong Y, Hong SH, Lee SJ,
Ryu DR, Lee W, Cho YH, Lee S, Ryu YJ, et al: RNA sequencing
identifies novel markers of non-small cell lung cancer. Lung
Cancer. 84:229–235. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Bisognin A, Pizzini S, Perilli L, Esposito
G, Mocellin S, Nitti D, Zanovello P, Bortoluzzi S and Mandruzzato
S: An integrative framework identifies alternative splicing events
in colorectal cancer development. Mol Oncol. 8:129–141. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Durrans A, Gao D, Gupta R, Fischer KR,
Choi H, El Rayes T, Ryu S, Nasar A, Spinelli CF, Andrews W, et al:
Identification of reprogrammed myeloid cell transcriptomes in
NSCLC. PLoS One. 10:e01291232015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Andrews S: FastQC: A quality control tool
for high throughput sequence data. http://www.bioinformatics.babraham.ac.uk/projects/fastqc/January
10–2016
|
|
14
|
Trapnell C, Roberts A, Goff L, Pertea G,
Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL and Pachter L:
Differential gene and transcript expression analysis of RNA-seq
experiments with TopHat and Cufflinks. Nat Protoc. 7:562–578. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Shen S, Park JW, Lu ZX, Lin L, Henry MD,
Wu YN, Zhou Q and Xing Y: rMATS: Robust and flexible detection of
differential alternative splicing from replicate RNA-Seq data. Proc
Natl Acad Sci USA. 111:pp. E5593–E5601. 2014; View Article : Google Scholar : PubMed/NCBI
|
|
16
|
da W Huang, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009.PubMed/NCBI
|
|
17
|
Sang Y, Bi X, Liu Y, Zhang W and Wang D:
Adverse prognostic impact of TGFB1 T869C polymorphism in
non-small-cell lung cancer. Onco Targets Ther. 10:1513–1518. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Han Y, Gao S, Muegge K, Zhang W and Zhou
B: Advanced applications of RNA sequencing and challenges.
Bioinform Biol Insights. 9 Suppl 1:29–46. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Krishnaswamy S, Mohammed AK, Amer OE,
Tripathi G, Alokail MS and Al-Daghri NM: Novel splicing variants of
recepteur d'origine nantais (RON) tyrosine kinase involving exons
15–19 in lung cancer. Lung Cancer. 92:41–46. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Fan YC, Min L, Chen H and Liu YL:
Alternative splicing isoform of T cell factor 4K suppresses the
proliferation and metastasis of non-small cell lung cancer cells.
Genet Mol Res. 14:14009–14018. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wang X, Xu X, Lu X, Zhang Y and Pan W:
Transcriptome Bioinformatical analysis of vertebrate stages of
Schistosoma japonicum reveals alternative splicing events. PLoS
One. 10:e01384702015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Hide WA, Babenko VN, van Heusden PA,
Seoighe C and Kelso JF: The contribution of exon-skipping events on
chromosome 22 to protein coding diversity. Genome Res.
11:1848–1853. 2001.PubMed/NCBI
|
|
23
|
Scagliotti GV and Novello S: The role of
the insulin-like growth factor signaling pathway in non-small cell
lung cancer and other solid tumors. Cancer Treat Rev. 38:292–302.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Sun Y, Zheng S, Torossian A, Speirs CK,
Schleicher S, Giacalone NJ, Carbone DP, Zhao Z and Lu B: Role of
insulin-like growth factor-1 signaling pathway in
cisplatin-resistant lung cancer cells. Int J Radiat Oncol Biol
Phys. 82:e563–e572. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Dávila D, Fernández S and Torres-Alemán I:
Astrocyte resilience to oxidative stress induced by insulin-like
growth factor I (IGF-I) involves preserved AKT (protein kinase B)
activity. J Biol Chem. 291:2510–2523. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Khawaja T, Chokshi A, Ji R, Kato TS, Xu K,
Zizola C, Wu C, Forman DE, Ota T, Kennel P, et al: Ventricular
assist device implantation improves skeletal muscle function,
oxidative capacity, and growth hormone/insulin-like growth factor-1
axis signaling in patients with advanced heart failure. J Cachexia
Sarcopenia Muscle. 5:297–305. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
De Palma M: The role of the immune system
in cancer: From mechanisms to clinical applications. Biochim
Biophys Acta. 1865:1–2. 2016.PubMed/NCBI
|
|
28
|
Soto-Ortiz L and Finley SD: A cancer
treatment based on synergy between anti-angiogenic and immune cell
therapies. J Theor Biol. 394:197–211. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Boros LG, D'Agostino DP, Katz HE, Roth JP,
Meuillet EJ and Somlyai G: Submolecular regulation of cell
transformation by deuterium depleting water exchange reactions in
the tricarboxylic acid substrate cycle. Med Hypotheses. 87:69–74.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Oosterveld LP, Allen JC Jr, Ng EY, Seah
SH, Tay KY, Au WL, Tan EK and Tan LC: Greater motor progression in
patients with Parkinson disease who carry LRRK2 risk variants.
Neurology. 85:1039–1042. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Marder K, Wang Y, Alcalay RN,
Mejia-Santana H, Tang MX, Lee A, Raymond D, Mirelman A,
Saunders-Pullman R, Clark L, et al: Age-specific penetrance of
LRRK2 G2019S in the Michael J. Fox Ashkenazi Jewish LRRK2
consortium. Neurology. 85:89–95. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Lee JW and Cannon JR: LRRK2 mutations and
neurotoxicant susceptibility. Exp Biol Med (Maywood). 240:752–759.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Bae JR and Lee BD: Function and
dysfunction of leucine-rich repeat kinase 2 (LRRK2): Parkinson's
disease and beyond. BMB Reports. 48:243–248. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Looyenga BD, Furge KA, Dykema KJ, Koeman
J, Swiatek PJ, Giordano TJ, West AB, Resau JH, Teh BT and MacKeigan
JP: Chromosomal amplification of leucine-rich repeat kinase-2
(LRRK2) is required for oncogenic MET signaling in papillary renal
and thyroid carcinomas. Proc Natl Acad Sci USA. 108:pp. 1439–1444.
2011; View Article : Google Scholar : PubMed/NCBI
|