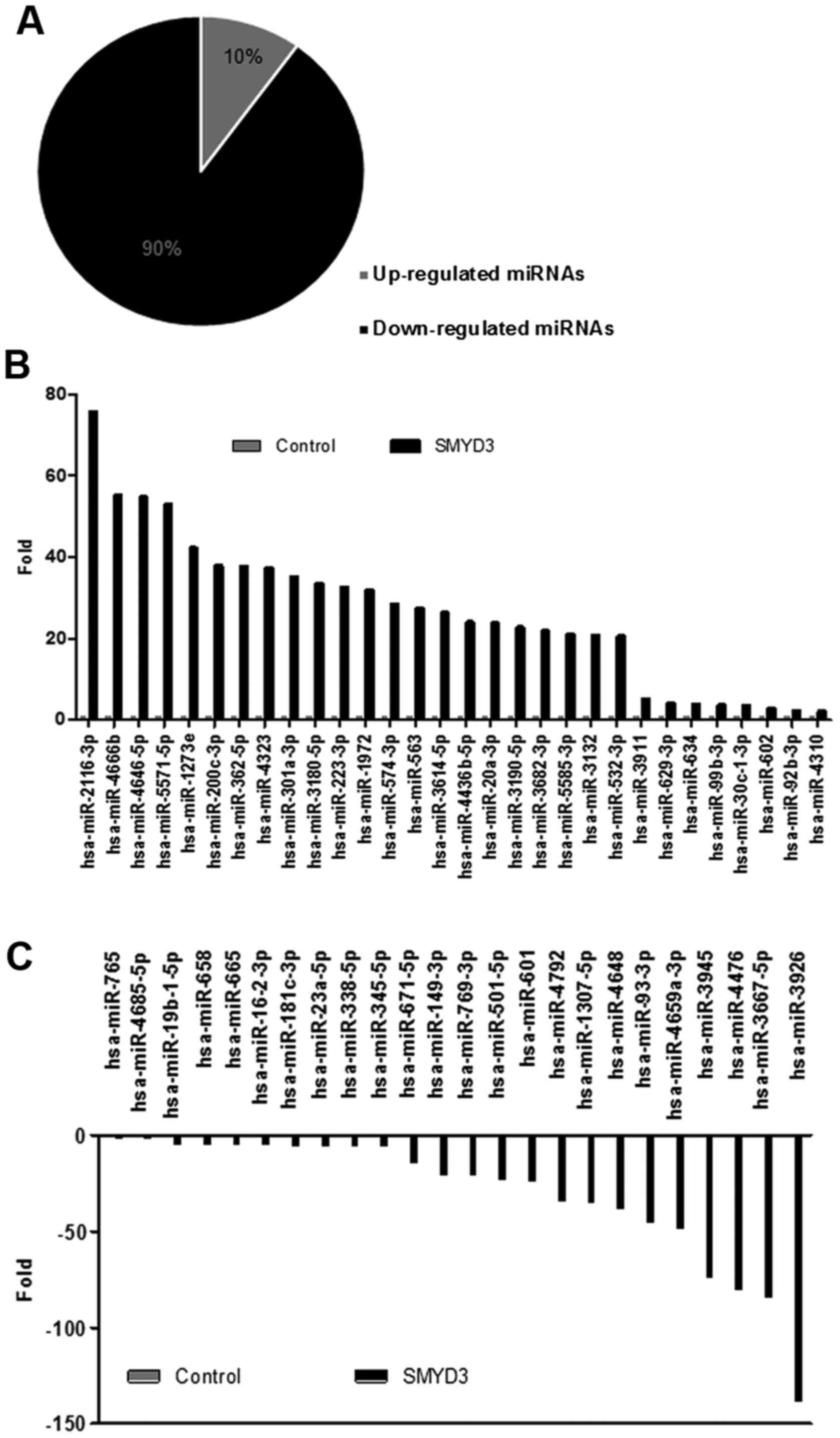
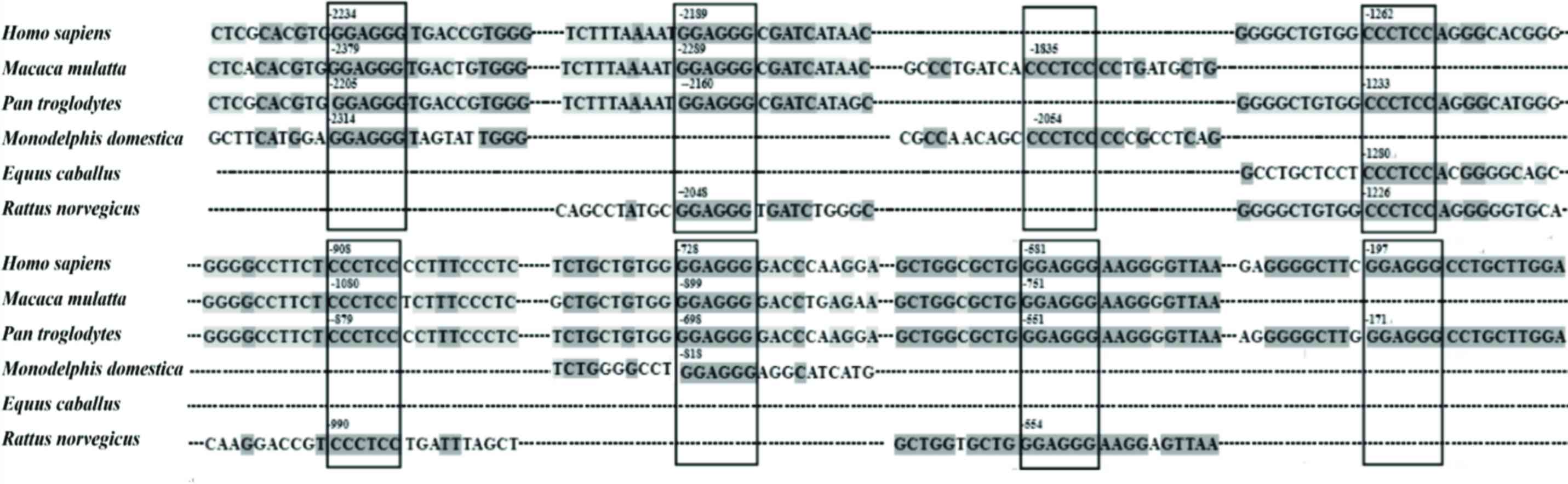
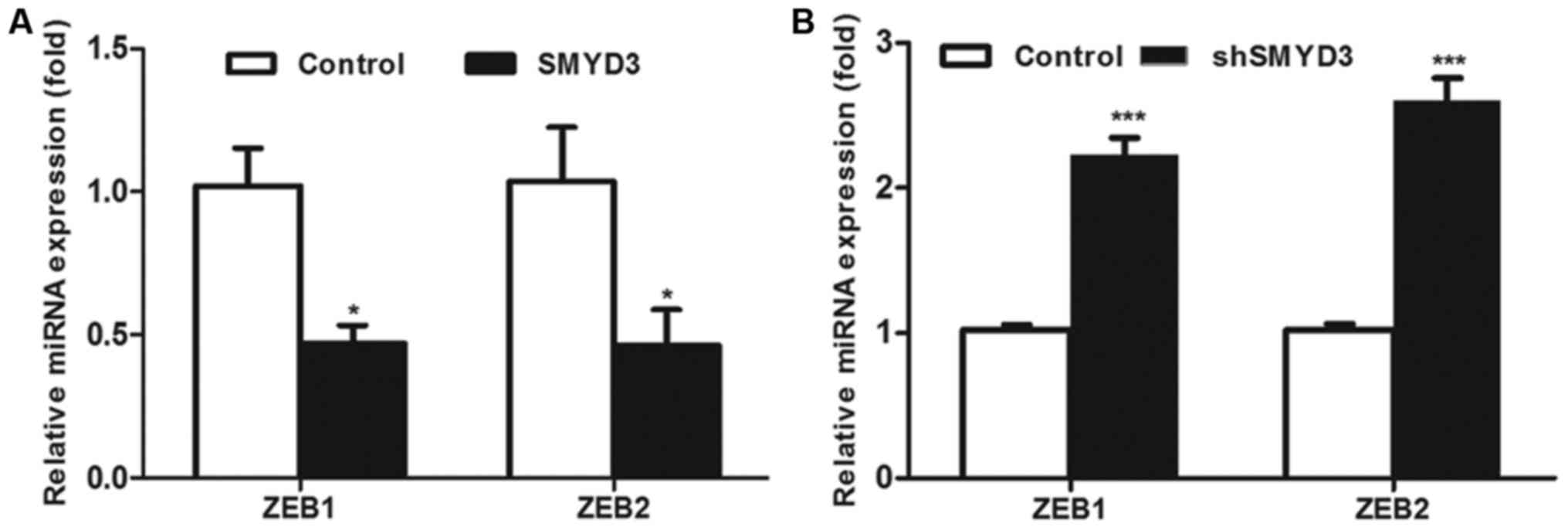
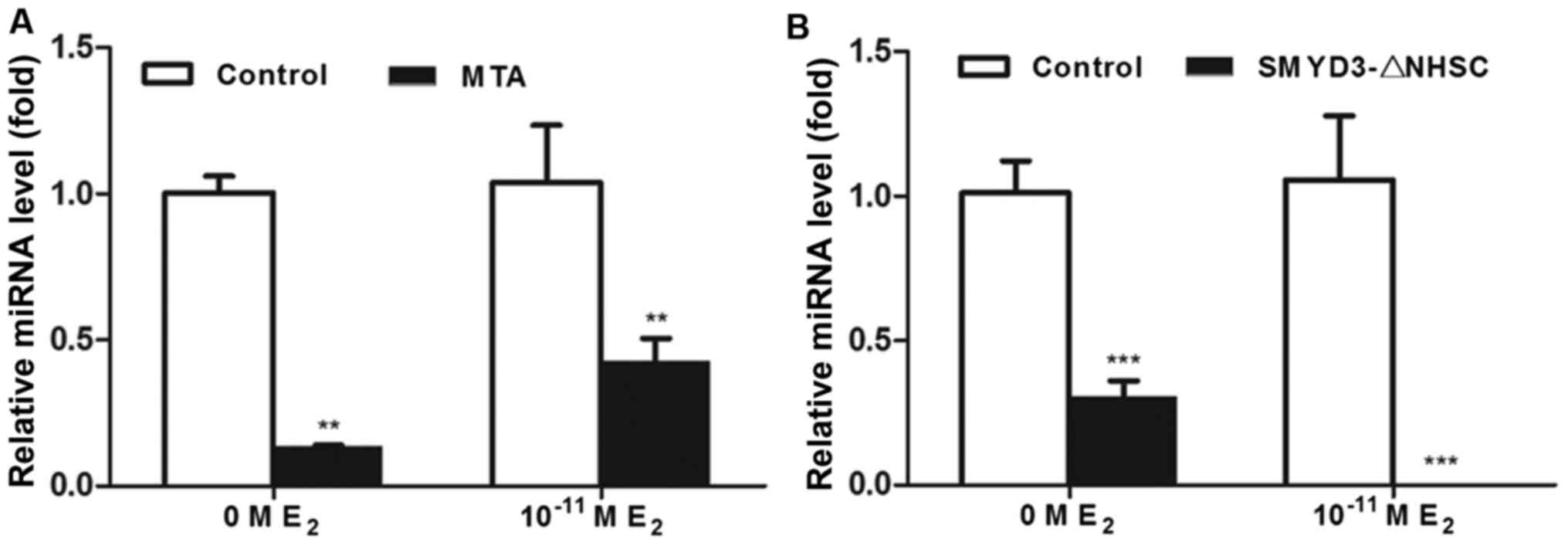
|
1
|
Hamamoto R, Furukawa Y, Morita M, Iimura
Y, Silva FP, Li M, Yagyu R and Nakamura Y: SMYD3 encodes a histone
methyltransferase involved in the proliferation of cancer cells.
Nat Cell Biol. 6:731–740. 2004. View
Article : Google Scholar : PubMed/NCBI
|
|
2
|
Hamamoto R, Silva FP, Tsuge M, Nishidate
T, Katagiri T, Nakamura Y and Furukawa Y: Enhanced SMYD3 expression
is essential for the growth of breast cancer cells. Cancer Sci.
97:113–118. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Wang SZ, Luo XG, Shen J, Zou JN, Lu YH and
Xi T: Knockdown of SMYD3 by RNA interference inhibits cervical
carcinoma cell growth and invasion in vitro. BMB Rep. 41:294–299.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Liu Y, Luo X, Deng J, Pan Y, Zhang L and
Liang H: SMYD3 overexpression was a risk factor in the biological
behavior and prognosis of gastric carcinoma. Tumour Biol.
36:2685–2694. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Van Aller GS, Reynoird N, Barbash O,
Huddleston M, Liu S, Zmoos AF, McDevitt P, Sinnamon R, Le B, Mas G,
et al: Smyd3 regulates cancer cell phenotypes and catalyzes histone
H4 lysine 5 methylation. Epigenetics. 7:340–343. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Foreman KW, Brown M, Park F, Emtage S,
Harriss J, Das C, Zhu L, Crew A, Arnold L, Shaaban S and Tucker P:
Structural and functional profiling of the human histone
methyltransferase SMYD3. PloS One. 6:e222902011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ambros V: The functions of animal
microRNAs. Nature. 431:350–355. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yahya SM and Elsayed GH: A summary for
molecular regulations of miRNAs in breast cancer. Clin Biochem.
48:388–396. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhang L, Deng L, Chen F, Yao Y, Wu B, Wei
L, Mo Q and Song Y: Inhibition of histone H3K79 methylation
selectively inhibits proliferation, self-renewal and metastatic
potential of breast cancer. Oncotarget. 5:10665–10677. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wang H, Ach RA and Curry B: Direct and
sensitive miRNA profiling from low-input total RNA. RNA.
13:151–159. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Hughes TR, Mao M, Jones AR, Burchard J,
Marton MJ, Shannon KW, Lefkowitz SM, Ziman M, Schelter JM, Meyer
MR, et al: Expression profiling using microarrays fabricated by an
ink-jet oligonucleotide synthesizer. Nature Biotechnol. 19:342–347.
2001. View Article : Google Scholar
|
|
12
|
Dennis G Jr, Sherman BT, Hosack DA, Yang
J, Gao W, Lane HC and Lempicki RA: DAVID: Database for annotation,
visualization and integrated discovery. Genome Biol. 4:P32003.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Katayama Y, Maeda M, Miyaguchi K, Nemoto
S, Yasen M, Tanaka S, Mizushima H, Fukuoka Y, Arii S and Tanaka H:
Identification of pathogenesis-related microRNAs in hepatocellular
carcinoma by expression profiling. Oncology Lett. 4:817–823.
2012.
|
|
15
|
Watahiki A and Wang Y, Morris J, Dennis K,
O'Dwyer HM, Gleave M, Gout PW and Wang Y: MicroRNAs associated with
metastatic prostate cancer. PloS One. 6:e249502011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Moossavi S and Rezaei N: Toll-like
receptor signalling and their therapeutic targeting in colorectal
cancer. Int Immunopharmacol. 16:199–209. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Place RF and Noonan EJ: Non-coding RNAs
turn up the heat: An emerging layer of novel regulators in the
mammalian heat shock response. Cell Stress Chaperones. 19:159–172.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Li J, Tan Q, Yan M, Liu L, Lin H, Zhao F,
Bao G, Kong H, Ge C, Zhang F, et al: miRNA-200c inhibits invasion
and metastasis of human non-small cell lung cancer by directly
targeting ubiquitin specific peptidase 25. Mol Cancer. 13:1662014.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Shi L, Fei X, Sun G, Wang Z, Wan Y, Zeng Y
and Guo J: Hypothermia stimulates glioma stem spheres to
spontaneously dedifferentiate adjacent non-stem glioma cells. Cell
Mol Neurobiol. 35:217–230. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Mycko MP, Cichalewska M, Machlanska A,
Cwiklinska H, Mariasiewicz M and Selmaj KW: MicroRNA-301a
regulation of a T-helper 17 immune response controls autoimmune
demyelination. Proc Natl Acad Sci USA. 109:pp. E1248–E1257. 2012;
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Archanioti P, Gazouli M, Theodoropoulos G,
Vaiopoulou A and Nikiteas N: Micro-RNAs as regulators and possible
diagnostic bio-markers in inflammatory bowel disease. J Crohns
Colitis. 5:520–524. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Li BS, Zhao YL, Guo G, Li W, Zhu ED, Luo
X, Mao XH, Zou QM, Yu PW, Zuo QF, et al: Plasma microRNAs, miR-223,
miR-21 and miR-218, as novel potential biomarkers for gastric
cancer detection. PLoS One. 7:e416292012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Madhavan D, Cuk K, Burwinkel B and Yang R:
Cancer diagnosis and prognosis decoded by blood-based circulating
microRNA signatures. Front Genet. 4:1162013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Guerit D, Philipot D, Chuchana P, Toupet
K, Brondello JM, Mathieu M, Jorgensen C and Noël D: Sox9-regulated
miRNA-574-3p inhibits chondrogenic differentiation of mesenchymal
stem cells. PLoS One. 8:e625822013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Cheung O, Puri P, Eicken C, Contos MJ,
Mirshahi F, Maher JW, Kellum JM, Min H, Luketic VA and Sanyal AJ:
Nonalcoholic steatohepatitis is associated with altered hepatic
MicroRNA expression. Hepatology. 48:1810–1820. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Marrale M, Albanese NN, Cali F and Romano
V: Assessing the impact of copy number variants on miRNA genes in
autism by Monte Carlo simulation. PLoS One. 9:e909472014.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Li X, Zhang Z, Yu M, Li L, Du G, Xiao W
and Yang H: Involvement of miR-20a in promoting gastric cancer
progression by targeting early growth response 2 (EGR2). Int J Mol
Sci. 14:16226–16239. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Fan X, Liu Y, Jiang J, Ma Z, Wu H, Liu T,
Liu M, Li X and Tang H: miR-20a promotes proliferation and invasion
by targeting APP in human ovarian cancer cells. Acta Biochim
Biophys Sin (Shanghai). 42:318–324. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ivashchenko A, Berillo O, Pyrkova A,
Niyazova R and Atambayeva S: The properties of binding sites of
miR-619-5p, miR-5095, miR-5096, and miR-5585-3p in the mRNAs of
human genes. Biomed Res Int. 2014:7207152014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Shin OS, Kumar M, Yanagihara R and Song
JW: Hantaviruses induce cell type- and viral species-specific host
microRNA expression signatures. Virology. 446:217–224. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Cheng Y, Kuang W, Hao Y, Zhang D, Lei M,
Du L, Jiao H, Zhang X and Wang F: Downregulation of miR-27a* and
miR-532-5p and upregulation of miR-146a and miR-155 in LPS-induced
RAW264.7 macrophage cells. Inflammation. 35:1308–1313. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Lu MC, Lai NS, Chen HC, Yu HC, Huang KY,
Tung CH, Huang HB and Yu CL: Decreased microRNA(miR)-145 and
increased miR-224 expression in T cells from patients with systemic
lupus erythematosus involved in lupus immunopathogenesis. Clin Exp
Immunol. 171:91–99. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Jeansonne D, Pacifici M, Lassak A, Reiss
K, Russo G, Zabaleta J and Peruzzi F: Differential effects of
microRNAs on glioblastoma growth and migration. Genes (Basel).
4:46–64. 2013.PubMed/NCBI
|
|
34
|
Jin Y, Tymen SD, Chen D, Fang ZJ, Zhao Y,
Dragas D, Dai Y, Marucha PT and Zhou X: MicroRNA-99 family targets
AKT/mTOR signaling pathway in dermal wound healing. PLoS One.
8:e644342013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Fang Y, Shen H, Cao Y, Li H, Qin R, Chen
Q, Long L, Zhu XL, Xie CJ and Xu WL: Involvement of miR-30c in
resistance to doxorubicin by regulating YWHAZ in breast cancer
cells. Braz J Med Biol Res. 47:60–69. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Sengupta S, Nie J, Wagner RJ, Yang C,
Stewart R and Thomson JA: MicroRNA 92b controls the G1/S checkpoint
gene p57 in human embryonic stem cells. Stem Cells. 27:1524–1528.
2009. View
Article : Google Scholar : PubMed/NCBI
|
|
37
|
Balakathiresan N, Bhomia M, Chandran R,
Chavko M, McCarron RM and Maheshwari RK: MicroRNA let-7i is a
promising serum biomarker for blast-induced traumatic brain injury.
J Neurotrauma. 29:1379–1387. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Kurokawa K, Tanahashi T, Iima T, Yamamoto
Y, Akaike Y, Nishida K, Masuda K, Kuwano Y, Murakami Y, Fukushima M
and Rokutan K: Role of miR-19b and its target mRNAs in
5-fluorouracil resistance in colon cancer cells. J Gastroenterol.
47:883–895. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Hackl M, Brunner S, Fortschegger K,
Schreiner C, Micutkova L, Mück C, Laschober GT, Lepperdinger G,
Sampson N, Berger P, et al: miR-17, miR-19b, miR-20a, and miR-106a
are down-regulated in human aging. Aging Cell. 9:291–296. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Sui W, Dai Y, Huang Y, Lan H, Yan Q and
Huang H: Microarray analysis of MicroRNA expression in acute
rejection after renal transplantation. Transpl Immunol. 19:81–85.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Tsuruta T, Kozaki K, Uesugi A, Furuta M,
Hirasawa A, Imoto I, Susumu N, Aoki D and Inazawa J: miR-152 is a
tumor suppressor microRNA that is silenced by DNA hypermethylation
in endometrial cancer. Cancer Res. 71:6450–6462. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Lezina L, Purmessur N, Antonov AV, Ivanova
T, Karpova E, Krishan K, Ivan M, Aksenova V, Tentler D, Garabadgiu
AV, et al: miR-16 and miR-26a target checkpoint kinases Wee1 and
Chk1 in response to p53 activation by genotoxic stress. Cell Death
Dis. 4:e9532013. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Ciafre SA, Galardi S, Mangiola A, Ferracin
M, Liu CG, Sabatino G, Negrini M, Maira G, Croce CM and Farace MG:
Extensive modulation of a set of microRNAs in primary glioblastoma.
Biochem Biophys Res Commun. 334:1351–1358. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Rathore MG, Saumet A, Rossi JF, de
Bettignies C, Tempé D, Lecellier CH and Villalba M: The NF-κB
member p65 controls glutamine metabolism through miR-23a. Int J
Biochem Cell Biol. 44:1448–1456. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Huang XH, Wang Q, Chen JS, Fu XH, Chen XL,
Chen LZ, Li W, Bi J, Zhang LJ, Fu Q, et al: Bead-based microarray
analysis of microRNA expression in hepatocellular carcinoma:
miR-338 is downregulated. Hepatol Res. 39:786–794. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Schou JV, Rossi S, Jensen BV, Nielsen DL,
Pfeiffer P, Høgdall E, Yilmaz M, Tejpar S, Delorenzi M, Kruhøffer M
and Johansen JS: miR-345 in metastatic colorectal cancer: A
non-invasive biomarker for clinical outcome in non-KRAS mutant
patients treated with 3rd line cetuximab and irinotecan. PloS One.
9:e998862014. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Rossi JJ: A novel nuclear miRNA mediated
modulation of a non-coding antisense RNA and its cognate sense
coding mRNA. EMBO J. 30:4340–4341. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Ke Y, Zhao W, Xiong J and Cao R: miR-149
inhibits non-small-cell lung cancer cells EMT by targeting FOXM1.
Biochem Res Int. 2013:5067312013. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Liu F, Xiong Y, Zhao Y, Tao L, Zhang Z,
Zhang H, Liu Y, Feng G, Li B, He L, et al: Identification of
aberrant microRNA expression pattern in pediatric gliomas by
microarray. Diagn Pathol. 8:1582013. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Sun Y, Wu J, Wu SH, Thakur A, Bollig A,
Huang Y and Liao DJ: Expression profile of microRNAs in c-Myc
induced mouse mammary tumors. Breast Cancer Res Treat. 118:185–196.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Ohdaira H, Nakagawa H and Yoshida K:
Profiling of molecular pathways regulated by microRNA 601. Comput
Biol Chem. 33:429–433. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Chang ST, Thomas MJ, Sova P, Green RR,
Palermo RE and Katze MG: Next-generation sequencing of small RNAs
from HIV-infected cells identifies phased microrna expression
patterns and candidate novel microRNAs differentially expressed
upon infection. MBio. 4:e00549–e00512. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Du L, Schageman JJ, Subauste MC, Saber B,
Hammond SM, Prudkin L, Wistuba II, Ji L, Roth JA, Minna JD and
Pertsemlidis A: miR-93, miR-98, and miR-197 regulate expression of
tumor suppressor gene FUS1. Mol Cancer Res. 7:1234–1243. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Alashti F Asghari and Minuchehr Z: MiRNAs
which target CD3 subunits could be potential biomarkers for
cancers. PLoS One. 8:e787902013. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Yang J, Liang Y, Han H and Qin H:
Identification of a miRNA signature in neutrophils after traumatic
injury. Acta Biochim Biophys Sin (Shanghai). 45:938–945. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Rustagi Y and Rani V: Screening of
microRNA as potential CardiomiRs in Rattus noveregicus heart
related dataset. Bioinformation. 9:919–922. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Rappa G, Mercapide J, Anzanello F, Pope RM
and Lorico A: Biochemical and biological characterization of
exosomes containing prominin-1/CD133. Mol Cancer. 12:622013.
View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Kim H, Heo K, Kim JH, Kim K, Choi J and An
W: Requirement of histone methyltransferase SMYD3 for estrogen
receptor-mediated transcription. J Biol Chem. 284:19867–19877.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Luo XG, Zhang CL, Zhao WW, Liu ZP, Liu L,
Mu A, Guo S, Wang N, Zhou H and Zhang TC: Histone methyltransferase
SMYD3 promotes MRTF-A-mediated transactivation of MYL9 and
migration of MCF-7 breast cancer cells. Cancer Lett. 344:129–137.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Jurmeister S, Baumann M, Balwierz A,
Keklikoglou I, Ward A, Uhlmann S, Zhang JD, Wiemann S and Sahin Ö:
MicroRNA-200c represses migration and invasion of breast cancer
cells by targeting actin-regulatory proteins FHOD1 and PPM1F. Mol
Cell Biol. 32:633–651. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Wu XD, Liu WL, Zeng K, Lei HY, Zhang QG,
Zhou SQ and Xu SY: Advanced glycation end products activate the
miRNA/RhoA/ROCK2 pathway in endothelial cells. Microcirculation.
21:178–186. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Cava C, Bertoli G and Castiglioni I:
Integrating genetics and epigenetics in breast cancer: Biological
insights, experimental, computational methods and therapeutic
potential. BMC Syst Biol. 9:622015. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Bischoff A, Huck B, Keller B, Strotbek M,
Schmid S, Boerries M, Busch H, Müller D and Olayioye MA: miR149
functions as a tumor suppressor by controlling breast epithelial
cell migration and invasion. Cancer Res. 74:5256–5265. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Zhang HF, Xu LY and Li EM: A family of
pleiotropically acting microRNAs in cancer progression, miR-200:
Potential cancer therapeutic targets. Curr Pharm Des. 20:1896–1903.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Sun Q, Liu T, Yuan Y, Guo Z, Xie G, Du S,
Lin X, Xu Z, Liu M, Wang W, et al: MiR-200c inhibits autophagy and
enhances radiosensitivity in breast cancer cells by targeting
UBQLN1. Int J Cancer. 136:1003–1012. 2015. View Article : Google Scholar : PubMed/NCBI
|