Introduction
Survivin, a member of the inhibitor of apoptosis
(IAP) gene family, has previously been reported as overexpressed in
numerous types of cancer and was associated with poor clinical
outcome (1–3). Survivin contains a baculovirus IAP
repeat, but does not contain a carboxyl-terminal RING finger. It is
prominently expressed in various cancer cell lines and in numerous
human types of cancer, including colon, lung and breast cancer
(1). The potential of survivin as a
biological target for anticancer therapies has been widely studied
(4–7).
The anticancer effects of survivin inhibition has been demonstrated
in melanoma cells (8), human colon
cancer cells (9) and K562 human
leukemia cell line (10).
Short interfering RNA (siRNA)-mediated gene
silencing is gradually becoming a powerful tool used to reduce
abnormally high expression for target genes, which results in its
potential applications in cancer therapy. Synthetic siRNA has been
used for targeting oncogenes and genes involved in various stages
of cancer cells, including proliferation, metastasis and apoptosis
(11,12). However, the broad applications of
siRNA in cancer therapy are based on a well-designed delivery
system that is able to efficiently deliver siRNA molecules into
tumors or target cells (13,14). Systemic therapeutic use of siRNA has
major limitations, including rapid degradation by nucleases and
renal clearance (15).
Nanocarriers are submicron size particles, ranging
from 1 to 1,000 nm in diameter (16).
They are able to overcome the majority of obstacles that limit the
therapeutic use of siRNA (17,18).
Nanoparticles are made of various biodegradable nanomaterials,
including liposomes, poylactic acid and polyethilenimine (19). The nanoliposomal siRNA carrier has
been demonstrated to efficiently carry and deliver siRNA in in
vivo systems (20,21). The present study aimed to investigate
the antitumor effect of survivin siRNA (si-survivin) delivered by
lipid nanoparticles. The results revealed that nanoliposomal
si-survivin may significantly reduce the expression level of
survivin and inhibit cell growth in vitro and in
vivo.
Materials and methods
Cell culture
LoVo, a human colon cancer cell line, was obtained
from Biomics Biotechnologies Co., Ltd. (Nantong, China) and
maintained in Dulbecco's modified Eagle's medium (Thermo Fisher
Scientific, Inc., Waltham, MA, USA) supplemented with 10% fetal
bovine serum (Hyclone; GE Healthcare, Logan, UT, USA). The medium
was supplemented with penicillin (100 U/ml) and streptomycin (100
mg/ml). The LoVo cells were incubated at 37°C in a humidified
atmosphere with 5% CO2.
Nanoliposomal siRNA construction
The siRNA oligonucleotides targeting human survivin
(si-survivin) were designed and synthesized by Biomics
Biotechnologies Co., Ltd. The sequence of si-survivin was as
follows: Sense, 5′-GCAUCUCUACAUUCAAGAA-3′ and anti-sense,
5′-UUCUUGAAUGUAGAGAUGC-3′. A scrambled sequence was used as the
negative control (si-NC) with the sequences as follows: Sense,
5′-UUCUCCGAACGUGUCACGU-3′ and anti-sense,
5′-ACGUGACACGUUCGGAGAA-3′. The siRNAs were then encapsulated into
disaturated phosphatidylcholine (DSPC; Avanti Polar Lipids,
Alablaster, AL, USA); Avanti Polar Lipids, Alablaster, AL, USA),
cholesterol, dioctadecyldimethylammonium chloride (DODAC) and
N-palmitoyl-sphingosine-1-succinyl (PEG-CerC16; Sigma-Aldrich;
Merck KGaA, Darmstadt, Germany) in 100% ethanol at a 25/45/25/2.5
molar ratio. The average diameter of nanoliposomal si-survivin and
si-NC was 70.7±29.077 and 64.9±26.128 nm, respectively.
In vitro transfection
Cultured LoVo cells were seeded at a density of
1×105 cells per well on a 24-well plate. Survivin and control
siRNA-liposomes were mixed with Opti-MEM (10 µl liposomes in a
total volume of 255 µl Opti-MEM; Life Technologies; Thermo Fisher
Scientific, Inc.) and left to stand for 5 min at room temperature.
Subsequently, nanoliposomes loaded with si-survivin or control
siRNA were added to each well at a concentration of 100 nmol/l at
37°C for 4–6 h. The mRNA level of survivin following transfection
was determined by reverse transcription-quantitative polymerase
chain reaction (RT-qPCR).
Total RNA isolation and RT-qRCR
Total RNA was isolated using RISO™RNA reagent
(Biomics Biotechnologies Co., Ltd.) and DNase (2.5 µl stock
solution diluted to a final volume of 100 µl; Biomics
Biotechnologies Co., Ltd.) was used for DNA digestion during the
extraction procedure. cDNA was synthesized using PrimeScript
reverse transcriptase (Takara Biotechnology Co., Ltd., Dalian,
China), according to the manufacturer's protocol. The mRNA level of
survivin was determined by qPCR using SensiMix™ One-Step
kit (Quantace, Taunton, MA, USA). RT-qPCR was performed on the ABI
PRISM Real-Time PCR system (Applied Biosystems; Thermo Fisher
Scientific, Inc., Waltham, MA, USA) using 2X One-Step qPCR mix
(12.5 µl), 50X SYBR-Green I (0.5 µl), primers (0.5 µl), and cDNA
template (100 ng in a total volume of 4 µl; all from Quantace;
Bioline USA, Inc., Taunton, MA, USA). The conditions for RT-qPCR
were as follows: 95°C for 10 min, followed by 40 cycles of 95°C for
20 sec, 58°C for 30 sec and 72°C for 30 sec. Specific primers were
used to detect survivin: Forward, 5′-ACGACCCCATAGAGGAACAT-3′ and
reverse, 5′-TCCGCAGTTTCCTCAAATTC-3′. The housekeeping gene GAPDH
was amplified as the internal control using specific primers:
Forward, 5′-GAAGGTGAAGGTCGGAGTC-3′ and reverse,
5′-GAAGATGGTGATGGGATTTC-3′. Relative gene expression was calculated
using the 2−ΔΔCq method (22). All analyses were performed in
triplicate.
Western blot analysis
To determine the protein expression level of
survivin following LoVo cellular uptake of the si-survivin complex,
total protein was extracted using lysis buffer supplemented with 25
mmol/l Tris-HCl, 150 mmol/l NaCl, 5 mmol/l EGTA, 5 mmol/l EDTA, 10
mmol/l NaF, 1 mmol/l phenylmethyl sulfonylfluoride, 1% TritonX-100,
0.5% Nonidet P40, 10 mg/l aprotinin and 10 mg/l leupeptin, as
previously described (10). Proteins
were quantified using a Bradford assay (Bio-Rad Laboratories, Inc.,
Hercules, CA, USA) according to the manufacturer's protocol, and
equal amounts of protein were subjected to 8% SDS-PAGE. The primary
antibodies used for western blotting were rabbit polyclonal
anti-survivin antibody (dilution, 1:500; cat. no. ab469; Abcam,
Cambridge, UK) and mouse anti-β-actin monoclonal antibody
(dilution, 1:400; cat. no. BM0005; Wuhan Boster Biological
Technology, Ltd., Wuhan, China) overnight at 4°C. The secondary
antibodies used were relative horseradish peroxidase-conjugated
secondary IgG antibodies (dilution, 1:500, cat. nos. BA1054 and
BA1051; Wuhan Boster Biological Technology, Ltd.) at room
temperature for 2 h. An enhanced chemiluminescence system
(Sino-American Biotechnology Co., Ltd., Luoyang, China) was used to
detect the expression levels of proteins. Band intensity
quantification was performed using Image J software version 1.441
(National Institutes of Health, Bethesda, MA, USA).
MTT assay
To detect the effect of si-survivin on the viability
of LoVo cells, MTT assay was performed using MTT reagent
(Sigma-Aldrich; Merck KGaA), according to the manufacture's
protocol. Briefly, 5×103 cells were seeded into 96-well plates
following overnight growth and were incubated with si-survivin
complex (100 nmol/l) or nanoliposomes with si-NC. MTT was added 48
h after uptake of siRNA and the cells were incubated at 37°C for an
additional 4 h. Absorbances were determined 4 h after the addition
of MTT and the optical density value at a wavelength of 570 nm was
determined.
In vivo tumorigenicity assay
Male athymic BALB/c nude mice (20–24 g, 7–9 weeks
old), were obtained from Shanghai Laboratory Animal Center
(Shanghai, China). Mice were maintained in specific pathogen-free
conditions with free access to food and water, under a constant
temperature of 22±2°C and a 12 h light/12 h dark cycle (7:00 a.m.
to 7:00 p.m.). All animal experiments were approved by the Ethics
Committee of the Capital Institute of Pediatrics (Beijing, China).
Cells were trypsinized, washed and re-suspended with sterile PBS. A
total of 200 µl cell suspension (6×106 cells) was
injected subcutaneously into the forelimb area of male BALB/c
athymic nude mice 4–6 weeks of age. When the xenograft tumors grew
to ~1 cm in diameter, the mice were sacrificed and the tumors were
obtained. The xenograft tumors were than sectioned into tissue
blocks (1×1×1 mm) and implanted into the right flank of forelimb
area of healthy male BALB/c athymic nude mice by subcutaneous
injection. The mice were then randomly divided into four groups (6
mice/group). Mice treated with nanolipsomal-si-NC complex at a
concentration of 3 mg/kg (twice a week for 5 weeks) were in the NC
group. Mice from the doxorubicin hydrochloride (DOX; Shenzhen Wanle
Pharmaceutical Co., Ltd., Wuhan, China). group were treated with an
intraperitoneal injection of 2.5 mg/kg (once a week for 5 weeks)
DOX as the positive control. Mice treated with
nanolipsomal-si-survivin intravenously (3 mg/kg, twice a week for 5
weeks) or intratumorally (50 µg per mouse, twice a week for 5
weeks) were the SU-IV or SU-IT groups, respectively. DOX was
administrated once a week, whereas siRNA-nanoliposomes were
administrated twice a week. All experimental procedures were
performed according to the guidelines of the Beijing Children's
Hospital, Capital Medicine University (Beijing, China). The present
study was approved by the Ethics Committee of The Beijing
Children's Hospital.
Tumor size was evaluated every 5 days using a
caliper and tumor volume was determined using the formula: Volume =
length × width × width/2. The length was the largest perpendicular
diameter and the width was the smallest. Following 33 days of
treatment, mice were sacrificed. Tumors were obtained, weighed and
stored at −80°C for further analysis. The relative tumor
proliferation rate, which was using to evaluate the effect of the
treatment, was calculated by the following formula:
Inhibitionrate(%)=[1–Tumorweight(treatment)Tumorweight(control)]x100
Immunohistochemistry (IHC) and
hematoxylin and eosin (H&E) staining
Tumor tissues were fixed in 4% paraformaldehyde at
4°C for at least 24 h and embedded in paraffin blocks to obtain
longitudinal and transverse sections. The sliced sections were then
used to perform IHC and H&E staining. IHC staining of samples
was performed as previously described (23) and the primary antibody used was the
rabbit polyclonal anti-survivin antibody (1:500, cat. no. ab76424,
Abcam). The sliced sections were stained with hematoxylin for 10
min followed by staining with eosin for 1–3 min at room
temperature, as described previously (24). Representative areas of digital
photomicrographs from each group were selected at a fixed
magnification of ×100 using a Nikon 50i light microscope (Nikon
Corporation, Tokyo, Japan).
Statistical analysis
Data is presented as the mean ± standard deviation.
Statistical significance was assessed using SPSS version 17.0 (IBM
SPSS, Armonk, NY, USA). Significance among groups was analyzed
using one-way analysis of variance, followed by Bonferroni's
post-hoc test to determine the differences between groups and
P<0.05 was considered to indicate a statistically significant
difference.
Results
Nanoliposomal si-survivin inhibits
cell proliferation in vitro
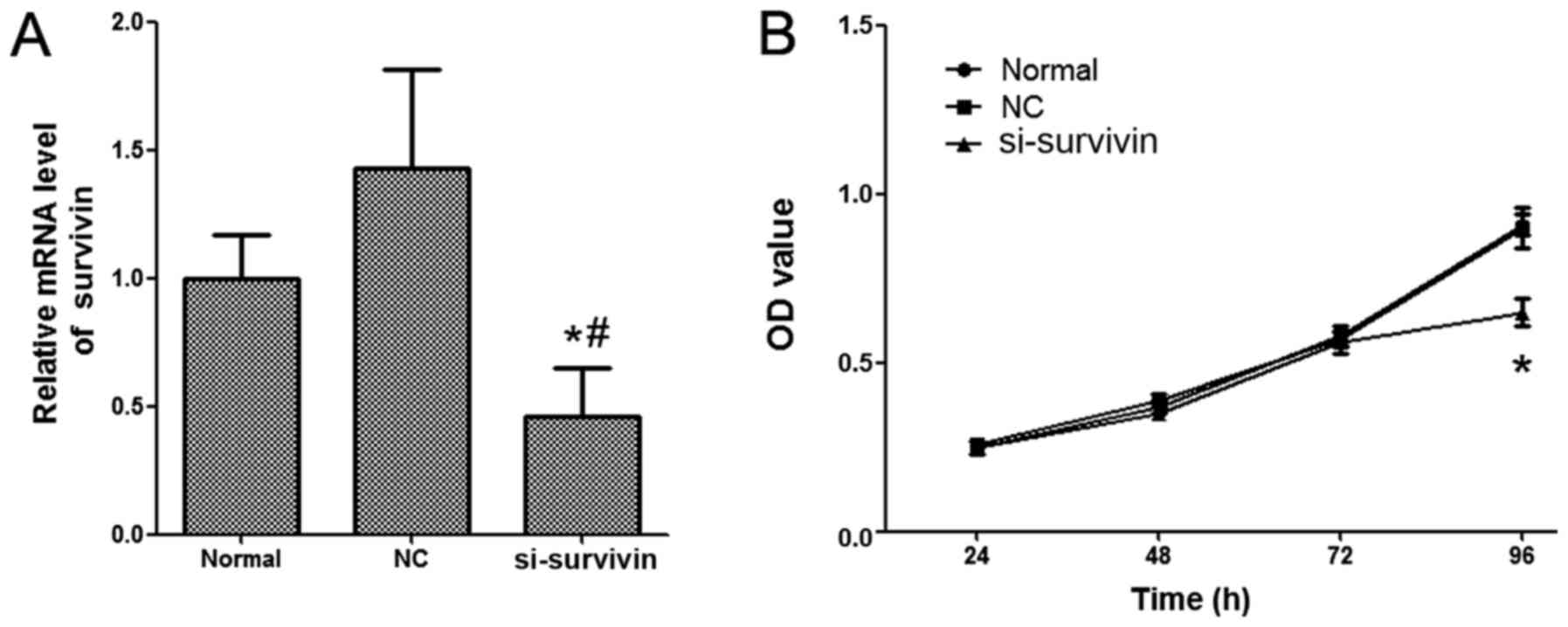
The expression levels of survivin in LoVo cells were
detected by RT-qPCR. As presented in Fig.
1A, the mRNA expression level of survivin was significantly
inhibited in the group treated with nanoliposomal si-survivin
compared with the normal and NC groups. In order to investigate the
effect of nanoliposomal si-survivin on the proliferation of colon
cancer, an MTT assay was performed. The results demonstrated that
the proliferation of LoVo cells was significantly inhibited by the
treatment of nanoliposomal si-survivin (Fig. 1B). No significant difference was
revealed in the nanoliposomal NC siRNA nanoliposome-transfected
cells and normal groups.
Nanoliposomal si-survivin inhibits
proliferation of colon cancer cells in vivo
The proliferation inhibition effect of nanoliposomal
si-survivin was further investigated in vivo. Tumor growth
was significantly reduced in the si-survivin nanoliposomes
treatment group compared with the NC group (P=0.031, SU-IV group
vs. NC group). The antitumor effect of DOX was greater compared
with the effect of nanoliposomal si-survivin. However, 3 mice in
the DOX group succumbed prior to the end of the experiment and the
average body weight in the DOX group was significantly lower
compared with in other groups, suggesting that DOX treatment may
have increased toxicity (Table I).
Mice treated with si-survivin nanoliposomes intratumorally or
intravenously, had similar body weights compared with mice in the
NC group (Table I). The tumor
inhibition rate of each treatment group is presented in Table I. The results revealed that the
inhibition rates of DOX treatment and intravenous administration of
nanoliposomal si-survivin were 68.9 and 31.1%, respectively;
whereas, intratumoral administration of si-survivin nanoliposomes
did not exhibit any obvious antitumor effects. Furthermore, tumor
growth was significantly inhibited with the treatment of
nanoliposomal si-survivin (Table
II).
 | Table I.Effect of antitumor activity of
survivin-targeted short interfering RNA nanoliposomes. |
Table I.
Effect of antitumor activity of
survivin-targeted short interfering RNA nanoliposomes.
|
| Mice, n | Body weight, g |
|
|
|---|
|
|
|
|
|
|
|---|
| Group | Pre-treatment | Post-treatment | Pre-treatment | Post-treatment | Tumor weight,
g | Inhibition rate,
% |
|---|
| NC-IV | 6 | 6 | 18.80±0.76 | 20.30±1.37 | 1.64±0.45 | / |
| DOX | 6 | 3 | 19.40±1.10 | 15.70±2.30 | 0.51±0.21 | 68.90 |
| SU-IT | 6 | 6 | 19.10±0.38 | 21.70±1.51 | 1.53±0.12 | 6.71 |
| SU-IV | 6 | 6 | 18.80±1.60 | 19.80±4.24 | 1.13±0.46 | 31.10 |
 | Table II.In vivo antitumor effects of
nanonanoliposomal survivin-targeted short interfering RNA on Balb/c
nude mice bearing LoVo tumor cells. |
Table II.
In vivo antitumor effects of
nanonanoliposomal survivin-targeted short interfering RNA on Balb/c
nude mice bearing LoVo tumor cells.
|
| NC-IV | DOX | SU-IT | SU-IV |
|---|
|
|
|
|
|
|
|---|
| Day | Tumor volume,
mm3 | RTV | T/C | Tumor volume,
mm3 | RTV | T/C, % | Tumor volume,
mm3 | RTV,
mm3 | T/C | Tumor volume,
mm3 | RTV | T/C |
|---|
| 0 | 27.65±8.38 | / | / | 18.60±6.36 | / | / | 24.01±5.26 | / | / | 28.57±4.56 | / | / |
| 1 | 69.40±22.48 | 2.53±0.39 | / | 48.68±22.94 | 2.58±0.57 | 102.03 | 57.48±4.02 | 2.46±0.40 | 97.29 | 73.86±13.63 | 2.58±0.25 | 102.13 |
| 5 | 132.35±48.63 | 4.30±1.56 | / | 98.45±59.13 | 4.09±2.89 | 95.27 | 94.11±18.01 | 3.98±0.71 | 92.73 | 105.26±28.28 | 3.69±0.85 | 85.82 |
| 8 | 198.39±70.56 | 6.30±2.95 | / | 131.38±72.69 | 5.51±3.42 | 87.48 | 148.22±28.50 | 6.38±1.74 | 101.31 | 187.36±79.46 | 6.39±1.95 | 101.47 |
| 12 | 311.80±129.29 | 9.97±5.57 | / | 185.71±96.64 | 7.87±4.53 | 78.90 | 232.60±57.12 | 9.80±2.28 | 98.27 | 269.46±115.52 | 9.24±3.04 | 92.68 |
| 15 | 478.16±182.52 | 13.83±6.35 | / | 165.61±107.26 | 8.34±6.43 | 60.29 | 372.07±59.58 | 15.94±3.84 | 115.25 | 380.30±123.96 | 13.19±3.18 | 95.34 |
| 19 | 674.53±269.54 | 21.49±13.10 | / | 271.88±175.04 | 13.82±10.73 | 64.32 | 524.02±109.30 | 22.41±5.92 | 104.29 | 401.46±175.41 | 14.03±4.43 | 65.31 |
| 22 | 789.29±114.64 | 25.76±15.80 | / | 290.86±201.10 | 15.07±12.55 | 58.49 | 641.16±169.76 | 27.08±6.33 | 105.12 | 498.24±184.16 | 17.43±4.97 | 67.69 |
| 26 |
1,009.91±154.20 | 37.65±15.40 | / | 454.00±250.80 | 23.49±17.20 | 62.40 | 764.20±120.19 | 32.79±7.53 | 87.10 | 655.62±189.66 | 22.94±8.31 | 60.10 |
| 29 |
1,133.79±283.40 | 42.40±16.90 | / | 380.59±59.83 | 27.33±21.73 | 64.45 | 941.79±107.78 | 40.66±9.86 | 95.89 | 784.17±158.16 | 27.45±7.14 | 64.73 |
| 33 |
1,452.67±314.40 | 54.20±24.90 | / | 645.41±225.20 | 34.96±23.83 | 64.51 |
1,121.84±102.95 | 48.43±10.91 | 89.36 | 889.65±197.29 | 31.13±7.06 | 57.45 |
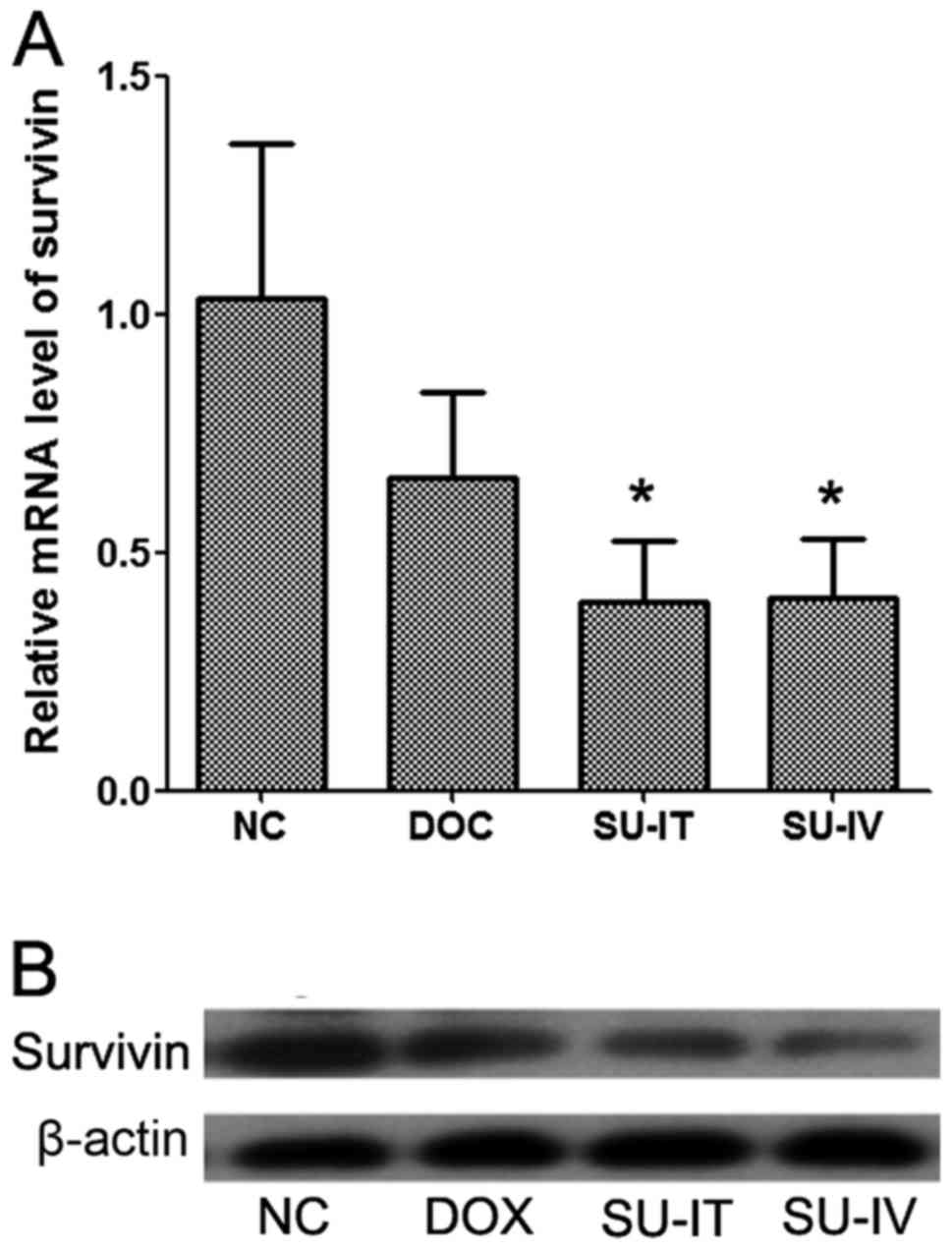
The mRNA and protein expression levels of survivin
in tumor tissues were detected by RT-qPCR and western blot
analysis, respectively. It was demonstrated that the mRNA and
protein expression levels of survivin were significantly reduced
following si-survivin treatment, intratumorally and intravenously,
compared with the NC group, which was treated with
nanolipsomal-si-NC complex. The expression levels of survivin were
decreased following DOX treatment but this was not significant
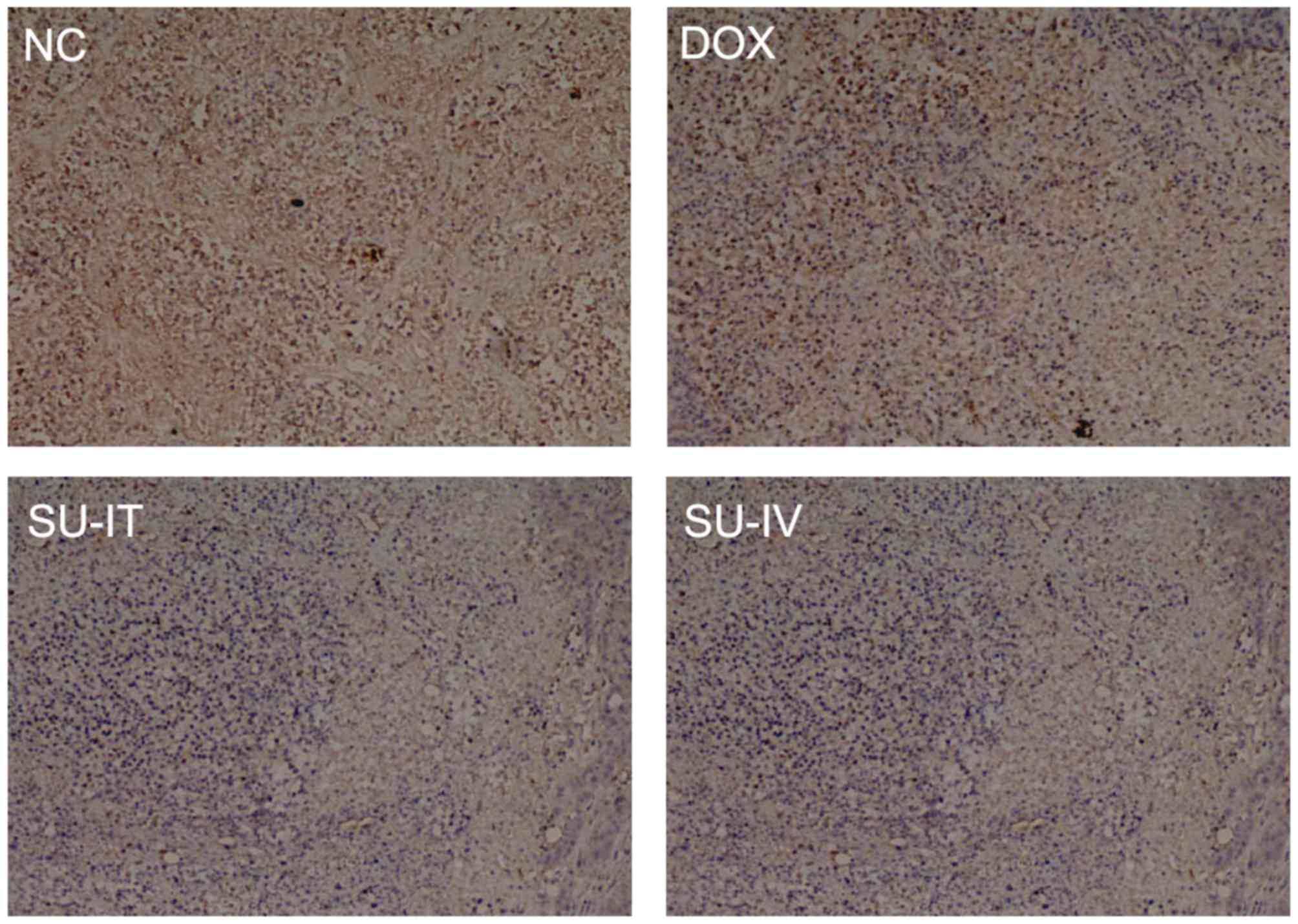
(Fig. 2). The results of IHC
indicated that survivin was mainly located in the cytoplasm and
that the expression level of survivin was reduced following
si-survivin treatment (Fig. 3). Tumor
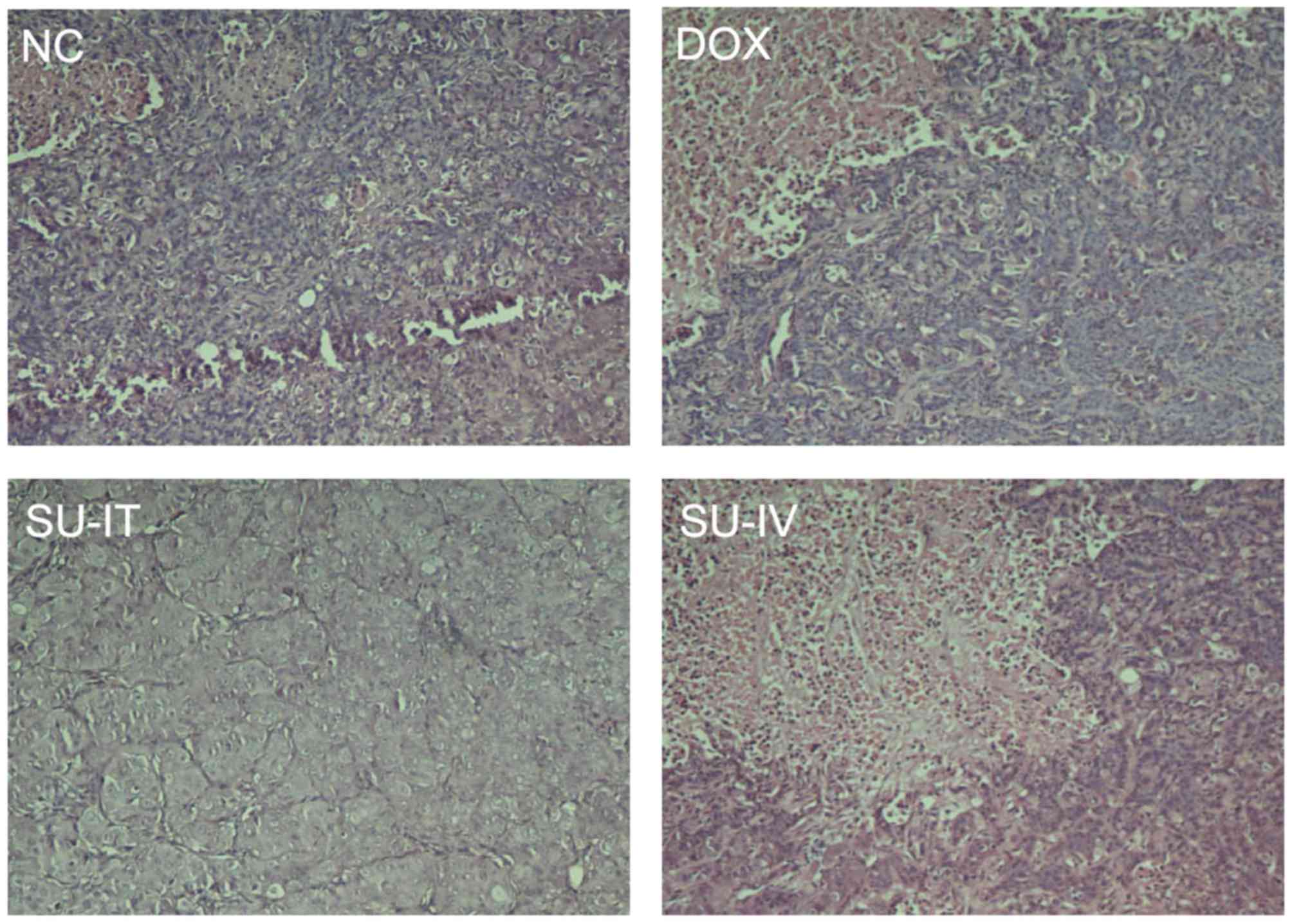
histological examination was detected by H&E staining and the
results are presented in Fig. 4. The
expression level of survivin was lower in the DOX group than in the
NC group (Fig. 4). These results
suggested that intravenous injection of nanoliposomal si-survivin
may significantly inhibit tumor growth in mice, and may be less
toxic compared with DOX treatment.
Discussion
Survivin serves an important role in cell apoptosis
and acts as a suppressor of apoptosis. It has been reported as
strongly expressed in numerous types of common human neoplasms, and
was associated with prognostic relevance, ionizing radiation and
cell resistance to antitumor agents (25). These findings suggest that survivin
may be a promising target for novel antitumor therapies. In recent
years, a number of various approaches have used to counteract
survivin to inhibit tumor growth or promote cell apoptosis
(26–28). It has been reported that survivin
anti-sense oligonucleotides may specifically inhibit the expression
levels of survivin mRNA and protein and reduce cell proliferation
in cell lines originated from various tumors, including lung, head,
neck and bladder cancer (29–31).
With the identification of RNA interference using
synthetic 21–23 nucleotide RNA duplexes, si-survivins have been
used for various types of cancer treatments (32,33).
Carvalho et al (34) reported
that si-survivins may specifically decrease the expression level of
survivin in HeLa cells and inhibited cell growth. This study also
demonstrated that si-survivins had a short half-life time and were
not detected 60 h following transfection (34). Paduano et al (23) revealed that si-survivins markedly
reduced the expression level of survivin and produced
supra-additive growth suppression in human androgen-independent
prostate cancer cells. Numerous previous studies have directly
added siRNA mimics into cell cultures (35–37).
However, the major limitations of direct addition of siRNA mimics
to cells are the instability and short half-life time. It has been
reported that the half-life of siRNA in serum was only ~15 min
(15).
In the present study, instead of using survivin
antisense oligonucleotide treatment or direct si-survivin
treatment, an alternative therapeutic approach for RNA interference
was used. si-survivins were encapsulated in the nanoliposomes and
then transfected into LoVo colon cancer cells. Nanocarriers have
been reported to be able to effectively deliver siRNAs and may also
prolong the half-lift time (17,18). Lipid
nanoparticles, which have been recognized as one of the most
efficient delivery systems for siRNAs, have been used extensively
(38,39). In the present study, lipid
nanoparticles were synthesized using DSPC, cholesterol, DODAC and
PEG-CerC16 at a 25/45/25/2.5 molar ratio. The particle diameter was
~70 nm following encapsulation with siRNAs. The nanoliposomal
siRNAs effectively delivered siRNAs into target cells. The results
of the present study demonstrated that the expression level of
survivin was significantly reduced and cell growth was
significantly inhibited following transfection with nanoliposomal
si-survivin in vitro. Furthermore, tumor growth was
significantly inhibited following systematic administration of
nanoliposomal si-survivin by intravenous injection into nude mice
with LoVo cell xenografts. Of note, the present study revealed that
the average body weight of mice following DOX treatment was lower
compared with other groups, whereas no significant changes of body
weight were observed in the group treated with si-survivin
nanoliposomes. A total of three mice succumbed prior to the end of
the experiment in the DOX treatment group. These results suggested
that lipid nanoparticles encapsulated with specific siRNAs may
effectively inhibit tumor growth with less toxicity compared with
traditional anticancer drugs.
In the present study, an efficient siRNA delivery
system using lipid nanoparticles was utilized to investigate the
potential treatment effect of si-survivin. The results demonstrated
that nanoliposomal si-survivin significantly reduced the expression
levels of survivin and inhibited cell growth in vitro.
Furthermore, si-survivin nanoliposomes significantly inhibited
tumor growth in nude mice bearing LoVo cell tumors with less
toxicity compared with DOX. The results of the present study
suggested that si-survivin delivered by nanoliposomes may be a
potential therapy for colon cancer treatment.
Acknowledgements
The present study was supported by the Chinese
National 863 Project (grant no. 2012AA020804).
Glossary
Abbreviations
Abbreviations:
|
siRNA
|
short interfering RNA
|
|
IAP
|
inhibitor of apoptosis
|
|
PLA
|
poylactic acid
|
|
PEI
|
polyethilenimine
|
|
MTT
|
methyl thiazolyl tetrazolium
|
|
IHC
|
immunohistochemistry
|
|
H&E
|
hematoxylin and eosin
|
References
|
1
|
Ambrosini G, Adida C and Altieri DC: A
novel anti-apoptosis gene, survivin, expressed in cancer and
lymphoma. Nat Med. 3:917–921. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Li F, Ambrosini G, Chu EY, Plescia J,
Tognin S, Marchisio PC and Altieri DC: Control of apoptosis and
mitotic spindle checkpoint by survivin. Nature. 396:580–584. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Sah NK, Khan Z, Khan GJ and Bisen PS:
Structural, functional and therapeutic biology of survivin. Cancer
Lett. 244:164–171. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zaffaroni N and Daidone MG: Survivin
expression and resistance to anticancer treatments: Perspectives
for new therapeutic interventions. Drug Resist Updat. 5:65–72.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Moriai R, Asanuma K, Kobayashi D, Yajima
T, Yagihashi A, Yamada M and Watanabe N: Quantitative analysis of
the anti-apoptotic gene survivin expression in malignant
haematopoietic cells. Anticancer Res. 21:595–600. 2001.PubMed/NCBI
|
|
6
|
Notarbartolo M, Cervello M, Dusonchet L,
Cusimano A and D'Alessandro N: Resistance to diverse apoptotic
triggers in multidrug resistant HL60 cells and its possible
relationship to the expression of P-glycoprotein, Fas and of the
novel anti-apoptosis factors IAP (inhibitory of apoptosis
proteins). Cancer Lett. 180:91–101. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Troeger A, Siepermann M, Escherich G,
Meisel R, Willers R, Gudowius S, Moritz T, Laws HJ, Hanenberg H,
Goebel U, et al: Survivin and its prognostic significance in
pediatric acute B-cell precursor lymphoblastic leukemia.
Haematologica. 92:1043–1050. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Moriyama M, Kano R, Maruyama H, Hasegawa A
and Kamata H: Small interfering RNA (siRNA) against the survivin
gene increases apoptosis in a canine melanoma cell line. J Vet Med
Sci. 72:1643–1646. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Cai M, Wang G, Tao K and Cai C: Induction
of apoptosis of human colon cancer cells by siRNA recombinant
expression vector targeting survivin gene. J Huazhong Univ Sci
Technolog Med Sci. 29:45–49. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Li J, Yue M, Shi X, Feng S, Tang R, Zhang
X, Liu R, Liu Z and Wang T: Evaluation of anti-cancer activity of
survivin siRNA delivered by folate receptor-targeted
polyethylene-glycol liposomes in K562-bearing xenograft mice.
Biomed Eng. 26:14500262014.
|
|
11
|
Hannon GJ and Rossi JJ: Unlocking the
potential of the human genome with RNA interference. Nature.
431:371–378. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Merritt WM, Lin YG, Han LY, Kamat AA,
Spannuth WA, Schmandt R, Urbauer D, Pennacchio LA, Cheng JF, Nick
AM, et al: Dicer, Drosha, and outcomes in patients with ovarian
cancer. N Engl J Med. 359:2641–2650. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Petros RA and DeSimone JM: Strategies in
the design of nanoparticles for therapeutic applications. Nat Rev
Drug Discov. 9:615–627. 2010. View
Article : Google Scholar : PubMed/NCBI
|
|
14
|
Burnett JC and Rossi JJ: RNA-based
therapeutics: Current progress and future prospects. Chem Biol.
19:60–71. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Tabernero J, Shapiro GI, LoRusso PM,
Cervantes A, Schwartz GK, Weiss GJ, Paz-Ares L, Cho DC, Infante JR,
Alsina M, et al: First-in-humans trial of an RNA interference
therapeutic targeting VEGF and KSP in cancer patients with liver
involvement. Cancer Discov. 3:406–417. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ozpolat B, Sood AK and Lopez-Berestein G:
Liposomal siRNA nanocarriers for cancer therapy. Adv Drug Deliv
Rev. 66:110–116. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ozpolat B, Sood AK and Lopez-Berestein G:
Nanomedicine based approaches for the delivery of siRNA in cancer.
J Intern Med. 267:44–53. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhou J, Shum KT, Burnett JC and Rossi JJ:
Nanoparticle-based delivery of RNAi therapeutics: Progress and
challenges. Pharmaceuticals (Basel). 6:85–107. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Allémann E, Rousseau J, Brasseur N,
Kudrevich SV, Lewis K and van Lier JE: Photodynamic therapy of
tumours with hexadecafluoro zinc phthalocyanine formulated in
PEG-coated poly (lactic acid) nanoparticles. Int J Cancer.
66:821–824. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yu B, Wang X, Zhou C, Teng L, Ren W, Yang
Z, Shih CH, Wang T, Lee RJ, Tang S and Lee LJ: Insight into
mechanisms of cellular uptake of lipid nanoparticles and
intracellular release of small RNAs. Pharm Res. 31:2685–2695. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wang Y, Xu Z, Guo S, Zhang L, Sharma A,
Robertson GP and Huang L: Intravenous delivery of siRNA targeting
CD47 effectively inhibits melanoma tumor growth and lung
metastasis. Mol Ther. 21:1919–1929. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Paduano F, Villa R, Pennati M, Folini M,
Binda M, Daidone MG and Zaffaroni N: Silencing of survivin gene by
small interfering RNAs produces supra-additive growth suppression
in combination with 17-allylamino-17-demethoxygeldanamycin in human
prostate cancer cells. Mol Cancer Ther. 5:179–186. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kingston EF, Goulding H and Bateman AC:
Vascular invasion is underrecognized in colorectal cancer using
conventional hematoxylin and eosin staining. Dis Colon Rectum.
50:1867–1872. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Shamsabadi FT, Eidgahi MR, Mehrbod P,
Daneshvar N, Allaudin ZN, Yamchi A and Shahbazi M: Survivin, a
promising gene for targeted cancer treatment. Asian Pac J Cancer
Prev. 17:3711–3719. 2016.PubMed/NCBI
|
|
26
|
Cheung CH, Huang CC, Tsai FY, Lee JY,
Cheng SM, Chang YC, Huang YC, Chen SH and Chang JY:
Survivin-biology and potential as a therapeutic target in oncology.
Onco Targets Ther. 6:1453–1462. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Mobahat M, Narendran A and Riabowol K:
Survivin as a preferential target for cancer therapy. Int J Mol
Sci. 15:2494–2516. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Chen XQ, Yang S, Kang MQ, Li ZY, Lu HS and
Lin TY: Survivin expression in human lung cancer and the influence
of its downregulation on the biological behavior of human lung
cancer cells. Exp Ther Med. 3:1010–1014. 2012.PubMed/NCBI
|
|
29
|
Cao C, Mu Y, Hallahan DE and Lu B: XIAP
and survivin as therapeutic targets for radiation sensitization in
preclinical models of lung cancer. Oncogene. 23:7047–7052. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Sharma H, Sen S, Lo Muzio L, Mariggiò A
and Singh N: Antisense-mediated downregulation of anti-apoptotic
proteins induces apoptosis and sensitizes head and neck squamous
cell carcinoma cells to chemotherapy. Cancer Biol Ther. 4:720–727.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Fuessel S, Kueppers B, Ning S, Kotzsch M,
Kraemer K, Schmidt U, Meye A and Wirth MP: Systematic in vitro
evaluation of survivin directed antisense oligodeoxynucleotides in
bladder cancer cells. J Urol. 171:2471–2476. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Li Z, Yang S, Chang T, Cao X, Shi L and
Fang G: Anti-angiogenesis and anticancer effects of a plasmid
expressing both ENDO-VEGI151 and small interfering RNA against
survivin. Int J Mol Med. 29:485–490. 2012.PubMed/NCBI
|
|
33
|
Li Z, Yin PH, Yang SS, Li QY, Chang T,
Fang L, Shi LX and Fang GE: Recombinant attenuated Salmonella
typhimurium carrying a plasmid co-expressing ENDO-VEGI151 and
survivin siRNA inhibits the growth of breast cancer in vivo. Mol
Med Rep. 7:1215–1222. 2013.PubMed/NCBI
|
|
34
|
Carvalho A, Carmena M, Sambade C, Earnshaw
WC and Wheatley SP: Survivin is required for stable checkpoint
activation in taxol-treated HeLa cells. J Cell Sci. 116:2987–2998.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Yang Z, Xie J, Zhu J, Kang C, Chiang C,
Wang X, Wang X, Kuang T, Chen F, Chen Z, et al: Functional
exosome-mimic for delivery of siRNA to cancer: In vitro and in vivo
evaluation. J Control Release. 243:160–171. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Bonnet S, Archer SL, Allalunis-Turner J,
Haromy A, Beaulieu C, Thompson R, Lee CT, Lopaschuk GD, Puttagunta
L, Bonnet S, et al: A mitochondria-K+ channel axis is suppressed in
cancer and its normalization promotes apoptosis and inhibits cancer
growth. Cancer Cell. 11:37–51. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Siu RW, Fragkoudis R, Simmonds P, Donald
CL, Chase-Topping ME, Barry G, Attarzadeh-Yazdi G, Rodriguez-Andres
J, Nash AA, Merits A, et al: Antiviral RNA interference responses
induced by Semliki Forest virus infection of mosquito cells:
Characterization, origin and frequency-dependent functions of
virus-derived small interfering RNAs. J Virol. 85:2907–2917. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Semple SC, Akinc A, Chen J, Sandhu AP, Mui
BL, Cho CK, Sah DW, Stebbing D, Crosley EJ, Yaworski E, et al:
Rational design of cationic lipids for siRNA delivery. Nat
Biotechnol. 28:172–176. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Yu B, Hsu SH, Zhou C, Wang X, Terp MC, Wu
Y, Teng L, Mao Y, Wang F, Xue W, et al: Lipid nanoparticles for
hepatic delivery of small interfering RNA. Biomaterials.
33:5924–5934. 2012. View Article : Google Scholar : PubMed/NCBI
|