Introduction
Hepatocellular carcinoma (HCC), the most frequent
subtype of primary liver cancer, accounting for between 85 and 90%
of liver cancer cases, is currently the third leading cause of
cancer-associated mortality worldwide (1). In the USA, 35,660 new HCC cases and
24,550 resulting mortalities were recorded in 2015 (2). HCC is prevalent in China and the country
accounts for >50% of all reported cases (3). A number of risk factors have been
implicated in HCC carcinogenesis and progression, including
alcoholic liver disease, non-alcoholic fatty liver disease, liver
cirrhosis, and hepatitis B and C infection (4–6). Despite
advances in HCC treatments, including hepatectomy and liver
transplantation, the rapid development of HCC results in poor
patient prognosis and a five-year survival rate of <5% (7,8). The
primary challenges in the diagnosis and treatment of HCC include,
poor early stage detection, distal metastasis and intrahepatic
recurrence following surgery (9). An
improved understanding of the underlying molecular mechanisms of
HCC initiation and progression may provide novel effective
therapeutic targets for the treatment of HCC, and improve
prognosis.
MicroRNAs (miRNAs) represent a group of highly
conserved, short non-coding RNA molecules of ~22 nucleotides in
length (10). miRNAs negatively
modulate protein expression through imperfect complementary
sequence pairing to the 3′ untranslated regions (3′-UTRs) of their
target mRNAs, leading to subsequent mRNA degradation or
translational repression (11,12). A
total of >1000 miRNAs have been identified in mammals; however,
the biological role of miRNA in carcinogenesis and cancer
progression remains unclear (13,14).
Increasing evidence suggests that abnormal miRNA expression
contributes to alterations in numerous cancer-associated processes,
including proliferation, cell cycle progression, apoptosis,
survival, migration, invasion and metastasis (15,16). The
dysregulation of miRNAs has been reported in various types of human
cancer, including human HCC, using miRNA detection systems
(17–19). These miRNAs may function as tumour
suppressors or oncogenes, and thus may be efficient prognostic
biomarkers of HCC tumorigenesis and progression (20,21).
miRNAs may also become effective targets in HCC therapy.
The expression and function of miRNA-708 (miR-708)
has been studied in several types of human cancer (22,23). The
present study investigated the expression patterns, biological
roles and underlying mechanisms of miR-708 in HCC. miR-708 was
observed to be significantly downregulated in HCC tissue samples
and cell lines. Reduced miR-708 expression was significantly
associated with increased HCC tumour stage. Furthermore, ectopic
miR-708 expression led to decreased cell proliferation, migration
and invasion through the direct targeting of SMAD family member 3
(SMAD3).
Materials and methods
HCC tissues and ethics statement
HCC and adjacent wild-type tissue sample pairs were
obtained from 108 patients with HCC who underwent a hepatectomy at
Yan'an City People's Hospital (Yanan, China). None of these
patients were treated with neoadjuvant radiotherapy or adjuvant
chemotherapy prior to surgery. All tissue specimens were
immediately frozen in liquid nitrogen following surgery and stored
at −80°C. The present study was approved by the Ethics Committee of
Yan'an City People's Hospital and informed consent was obtained
from all subjects.
Cell lines and cell culture
The human HCC cell lines, HepG2 and SMMC-7721, and
the wild-type hepatic cell line L02, were purchased from the Cell
Type Culture Collection of the Chinese Academy of Sciences
(Shanghai, China). Cells were cultured in Dulbecco's Modified
Eagle's medium (DMEM; Gibco; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA) containing 10% heat-inactivated foetal bovine
serum (FBS; Gibco; Thermo Fisher Scientific, Inc.) at 37°C in a
humidified atmosphere containing 5% CO2.
Cell transfection
A mature miR-708 mimic and scrambled miRNA mimic
negative control (miR-NC) were purchased from Shanghai GenePharma
Co., Ltd. (Shanghai, China). The sequence of the miR-708 mimic was
5′-AAGGAGCUUACAAUCUAGCUGGG-3′. The sequence of the miR-NC mimic was
5′-UUCUCCGAACGUGUCACGUTT-3′. Cells were seeded into six-well plates
at between 60 and 70% confluence and transfected with the miRNA
mimics using Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) reagent according to the manufacturer's
protocol.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR) analysis
Total RNA was extracted from homogenised tissue
samples and cell lines using TRIzol® reagent (Invitrogen; Thermo
Fisher Scientific, Inc.). cDNA was synthesised using a TaqMan®
MicroRNA Reverse Transcription kit (Applied Biosystems; Thermo
Fisher Scientific, Inc.) according to the manufacturer's protocol.
qPCR was performed using the TaqMan MicroRNA Assay kit (Applied
Biosystems; Thermo Fisher Scientific, Inc.) and an Applied
Biosystems® 7900HT Fast Real-Time PCR system (Thermo Fisher
Scientific, Inc.). The sequences of the primers used were as
follows: miR-708 forward, 5′-CGGCGGAAGGAGCTTACAATCTA-3′ and
reverse, 5′-GTGCAGGGTCCGAGG-3′; and U6 forward,
5′-CTCGCTTCGGCAGCACATATACT-3′ and reverse,
5′-ACGCTTCACGAATTTGCGTGTC-3′. The thermocycling conditions were as
follows: 95°C for 10 min; 40 cycles of denaturation at 95°C for 15
sec and annealing at 60°C for 1 min; followed by a final elongation
step at 72°C for 10 min. U6 spliceosomal RNA was used as the
endogenous control. Fold changes in relative expression were
calculated using the 2−ΔΔCq method (24).
Cell proliferation assay
The Cell Counting Kit-8 (CCK-8; Dojindo Molecular
Technologies, Inc., Kumamoto, Japan) assay was used to verify the
effect of miR-708 expression in HCC on cell proliferation. A total
of 3000 transfected cells (100 µl) were seeded into each well of a
96-well plate. The cells were subsequently incubated at 37°C for
24, 48, 72 and 96 h and at each time point the CCK-8 assay was
performed. A total of 10 µl CCK-8 reagent was added and incubated
at 37°C for an additional 2 h. The absorbance was determined at a
wavelength of 450 nm using a microplate reader (Thermo Fisher
Scientific, Inc.).
Cell migration and invasion
assays
Transwell® chambers (8 µm; Corning Incorporated,
Corning, NY, USA) and Matrigel® (BD Biosciences, Franklin Lakes,
NJ, USA)-coated Transwell chambers were inserted into 24-well
plates and used in cell migration and invasion assays,
respectively. A total of 1×105 transfected cells (300
µl) in FBS-free DMEM were added to the upper Transwell chamber. A
total of 500 µl DMEM containing 20% FBS was added to the lower
Transwell chamber. Cells were incubated at 37°C for 12 h (migration
assay) and 24 h (invasion assay). Subsequently, remaining cells in
the upper chamber were removed using a cotton-tipped swab.
Migratory and invasive cells in the lower chamber were fixed with
100% methanol, stained with 0.5% crystal violet (Sigma-Aldrich;
Merck Millipore, Darmstadt, Germany) and washed with PBS. Migratory
and invasive cells in five randomly selected visual fields were
counted using a light microscope.
Western blotting
Total protein was isolated from transfected cells 72
h following transfection using radioimmunoprecipitation assay
buffer (Beyotime Institute of Biotechnology, Haimen, China)
containing protease and phosphatase inhibitors. The protein
concentration was determined using a Bicinchoninic Acid Protein
Assay kit (Beyotime Institute of Biotechnology). Equal masses of
total protein (20 µg) were separated using SDS-PAGE on a 10% gel
(Beyotime Institute of Biotechnology) and transferred to
polyvinylidene difluoride membranes (EMD Millipore, Billerica, MA,
USA). Following blocking in 10% skimmed milk at room temperature
for 2 h, the membranes were incubated with rabbit anti-human
monoclonal SMAD3 antibody (1:1,000; cat. no. 9523s; Cell Signaling
Technology, Inc., Danvers, MA, USA) and mouse anti-human monoclonal
β-actin antibody (1:1,000; cat. no. sc-47778; Santa Cruz
Biotechnology, Inc., Dallas, TX, USA) at room temperature for 4 h.
The membranes were subsequently washed five times with TBS and
Tween 20 and incubated with an anti-rabbit (1:2,000; cat. no.
sc-2004; Santa Cruz Biotechnology, Inc.) or anti-mouse (1:2,000;
cat. no. sc-2005; Santa Cruz Biotechnology, Inc.) horseradish
peroxidase-conjugated secondary antibody for 2 h at room
temperature. Protein bands were detected using the Pierce™ ECL
Western Blotting Substrate (Pierce Biotechnology, Inc., Rockford,
IL, USA), and Quantity One® software (version 4.62; Bio-Rad
Laboratories, Inc., Hercules, CA, USA).
Dual luciferase reporter assay
Dual luciferase reporter assays were used to confirm
whether SMAD3 is a direct target of miR-708. HCC cells were seeded
into 24-well plates at a density of between 50 and 60% confluence,
and transfected with the miRNA mimics and plasmids containing the
wild-type SMAD3 3′-UTR (pMIR-SMAD3-3′UTR Wt) or a mutated SMAD3
3′-UTR (pMIR-SMAD3-3′UTR Mut; both GenePharma Co., Ltd.) using
Lipofectamine 2000. A total of 48 h following transfection, a
Dual-Luciferase Reporter Assay system (cat. no. E1910; Promega
Corporation, Madison, WI, USA) was used to determine firefly and
Renilla luciferase activities. Renilla luciferase
activity was used as the internal control.
Statistical and bioinformatic
analysis
The putative miR-708 target genes were predicted by
bioinformatic analysis using the miRanda database (www.microrna.org). Values are presented as the mean ±
standard deviation of triplicate data and were compared using the
Student's t-test. Statistical analysis was performed using SPSS
(version 17; SPSS, Inc., Chicago, IL, USA). P<0.05 was
considered to indicate a statistically significant difference.
Results
miR-708 is downregulated in HCC
tissues and cell lines
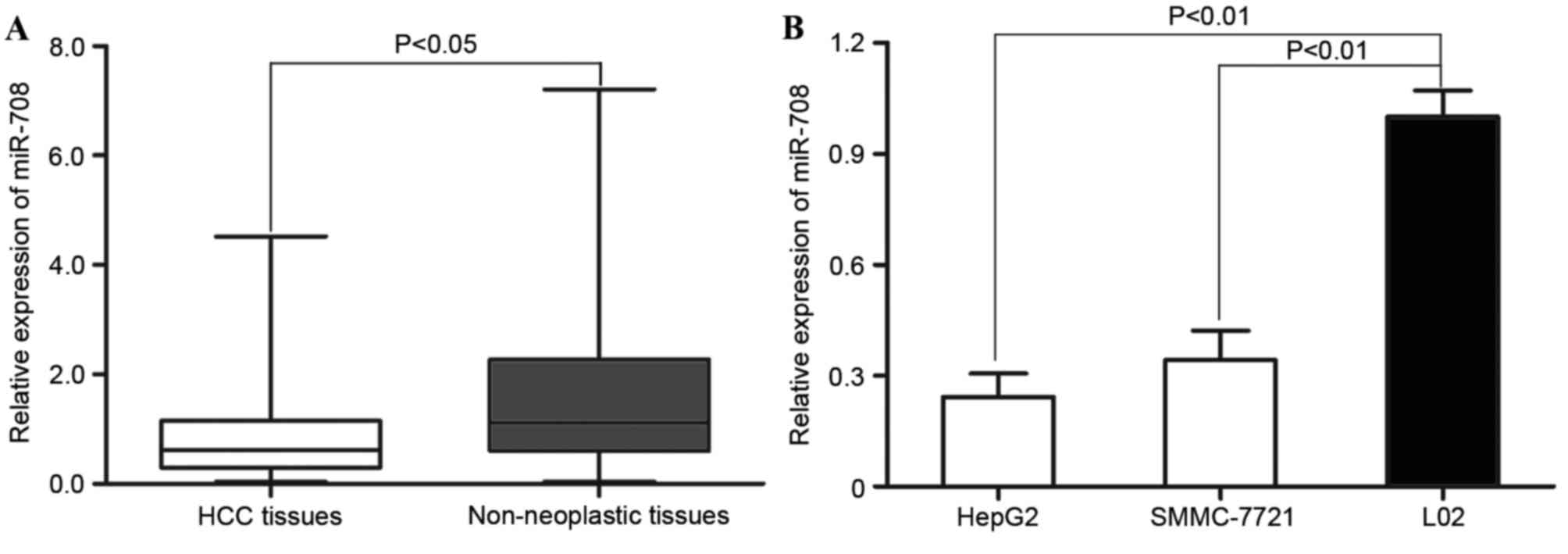
To evaluate the biological role of miR-708 in human
HCC, RT-qPCR analysis was performed to measure miR-708 expression
in HCC and adjacent wild-type tissue samples. As shown in Fig. 1A, miR-708 expression was significantly
decreased in HCC tissue samples compared with the adjacent
wild-type tissue samples (P=0.023). In addition, miR-708 expression
in the HCC cell lines HepG2 and SMMC-7721, and normal hepatic cell
line L02, was analysed. As shown in Fig.
1B, miR-708 was also significantly decreased in the HCC cell
lines compared with the wild-type cell line (HepG2, P=0.012;
SMMC-7721, P=0.017). These results suggest that miR-708 is involved
in the regulation of HCC malignancy.
Association between miR-708 expression
and HCC tumour stage
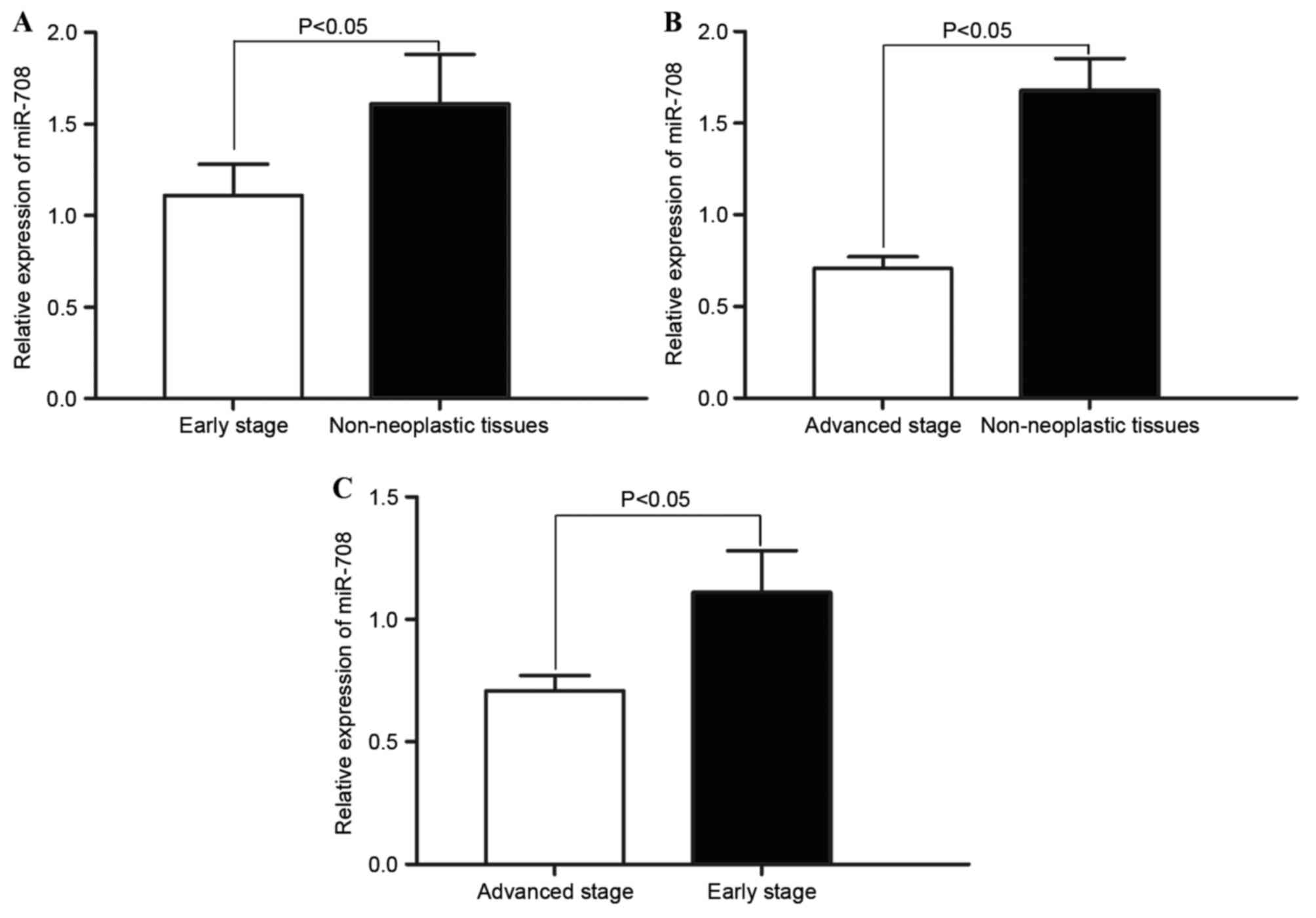
The association between decreased miR-708 expression
and HCC tumour stage was investigated. Statistical analysis
demonstrated that miR-708 expression in early (I–II) and advanced
(III–IV) tumour stages was significantly decreased compared with
the non-neoplastic tissues (P=0.034; Fig.
2A; P=0.026; Fig. 2B). In
addition, significantly decreased miR-708 expression was observed
in advanced stage HCC tissue samples compared with the early stage
HCC tissue samples (P=0.030; Fig.
2C). These results indicate that decreased miR-708 expression
level is associated with increased HCC tumour stage.
Increased miR-708 expression inhibits
HCC cell proliferation
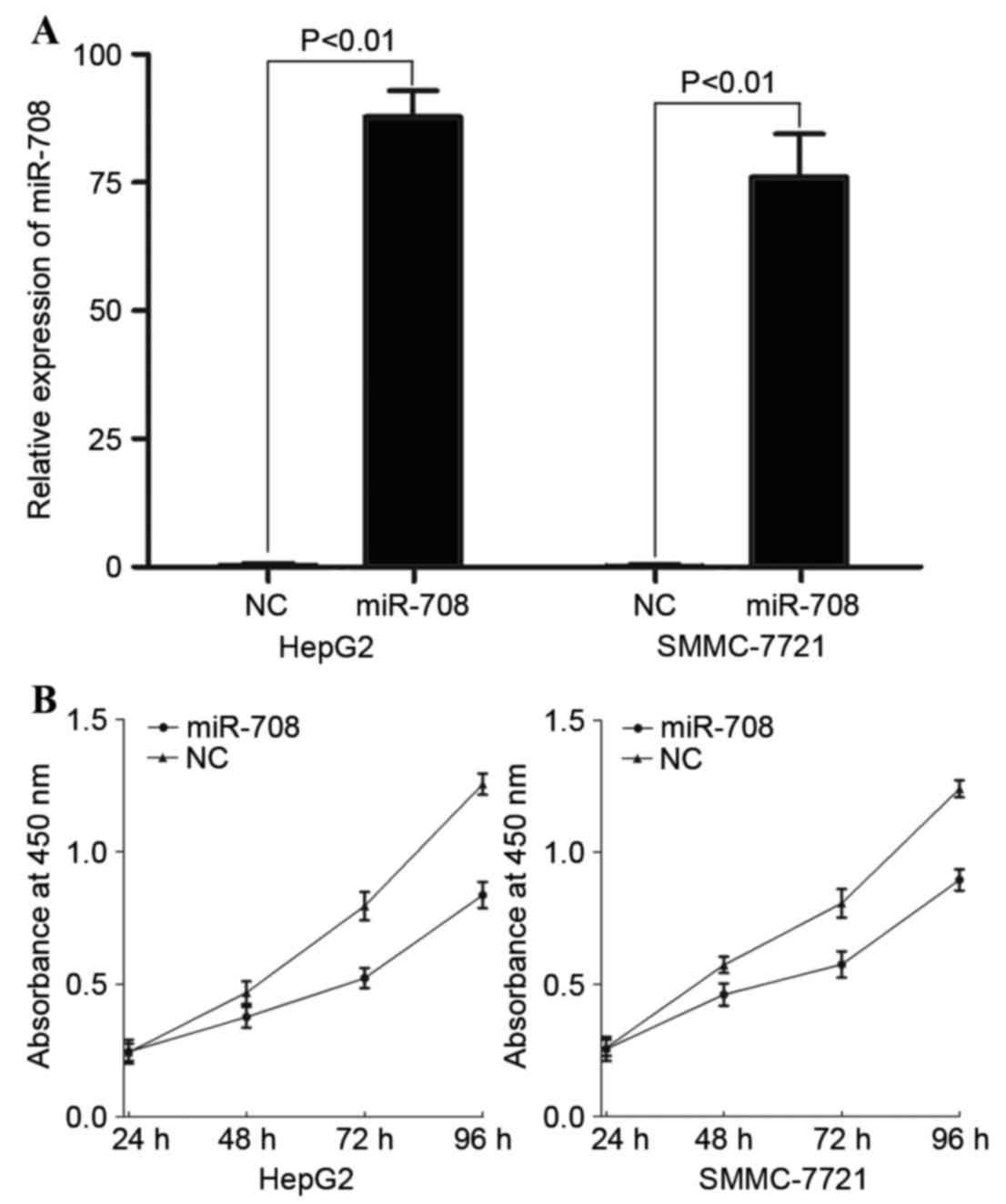
To investigate the role of miR-708 in HCC, a miR-708
mimic was transfected into HepG2 and SMMC-7721 cells. Following
transfection, RT-qPCR was performed to evaluate the transfection
efficiency, and miR-708 expression was observed to be significantly
increased in miR-708-transfected HepG2 (P=0.00000013) and SMMC-7721
(P=0.00000036) cells compared with the miR-NC-transfected cells
(Fig. 3A). Cell proliferation assays
were performed to assess the effect of miR-708 expression on cell
proliferation. As shown in Fig. 3B,
miR-708 overexpression inhibited cell proliferation in HepG2 and
SMMC-7721 cells compared with the miR-NC-transfected cells. A total
of 96 h following transfection, cell proliferation was inhibited by
33.24±3.5 and 27.83±2.7% in miR-708-transfected HepG2 (P=0.018) and
SMMC-7721 (P=0.021) cells, respectively.
miR-708 inhibits cell migration and
invasion of HCC
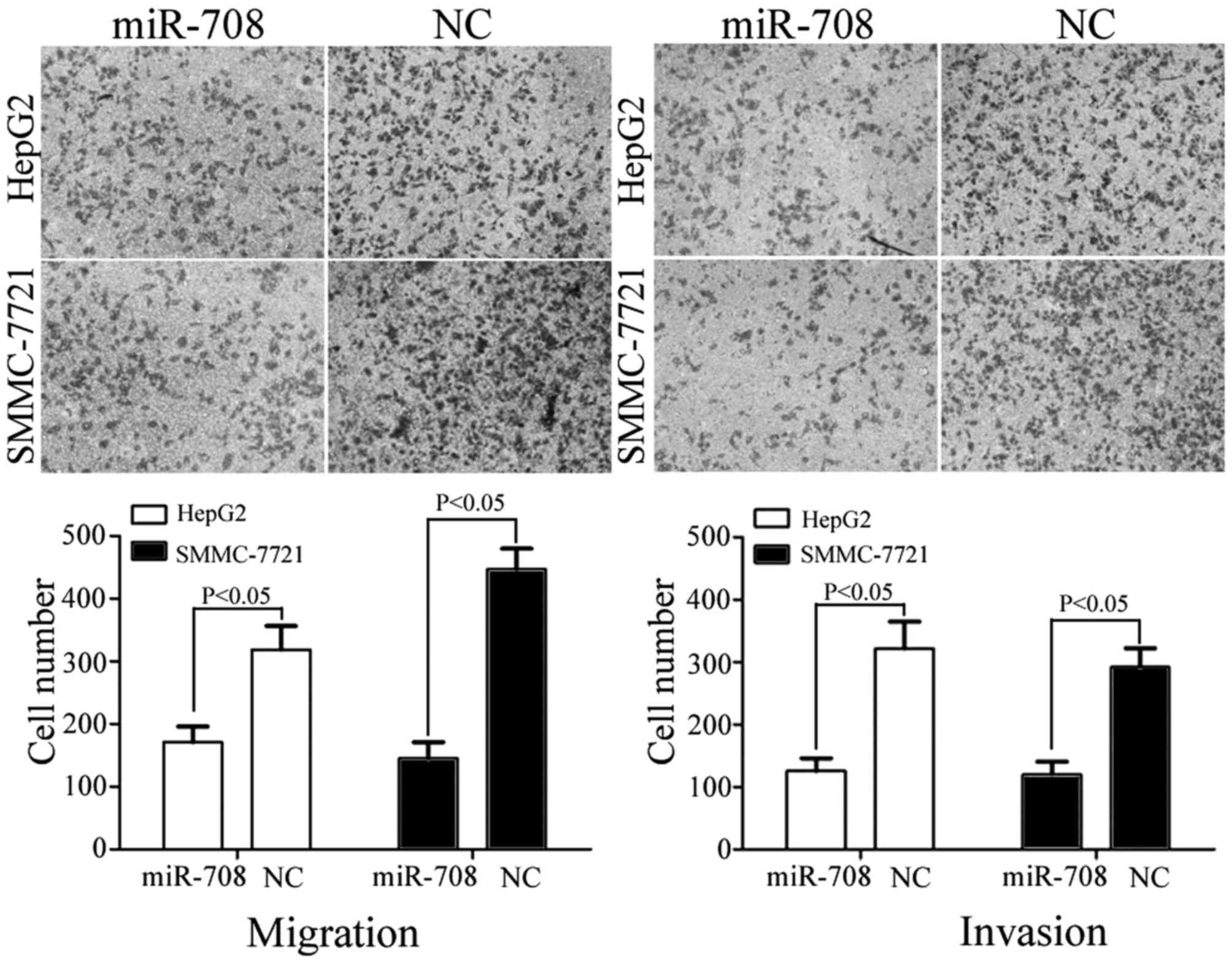
Cell migration and invasion assays were performed to
investigate the role of miR-708 in the regulation of metastasis in
HCC. As shown in Fig. 4, miR-708
overexpression resulted in the significant suppression of migration
in HepG2 (P=0.033) and SMMC-7721 (P=0.024) cells compared with the
miR-NC-transfected cells. In addition, miR-708 overexpression led
to a significant decrease in the number of invasive HepG2 (P=0.020)
and SMMC-7721 (P=0.026) cells compared with the miR-NC-transfected
cells (Fig. 4). These observations
suggest that miR-708 is a negative regulator of HCC metastasis.
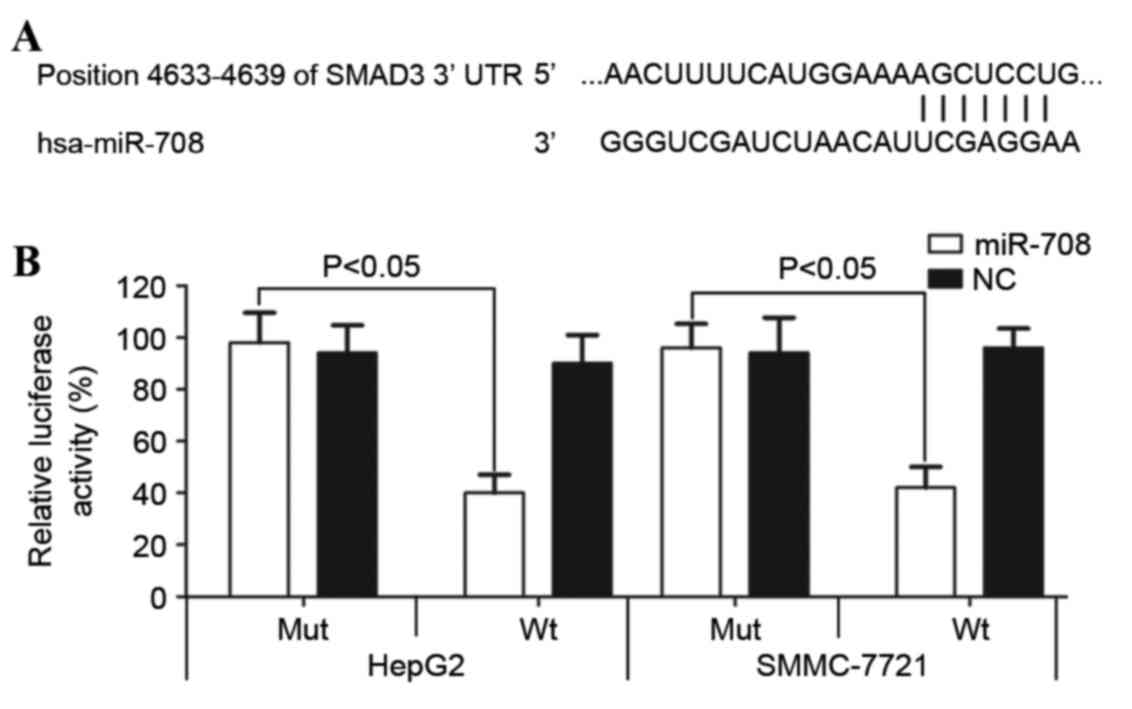
miR-708 directly targets the 3′-UTR of
SMAD3
To further understand the underlying mechanism of
miR-708, bioinformatic analysis was performed and a number of
candidate target genes were identified. SMAD3, an oncogene in
numerous types of cancer (25,26), was
selected for further study due to the putative miR-708 target
sequences contained within the SMAD3 3′-UTR (Fig. 5A). The dual luciferase reporter assay
was performed to evaluate the interaction between miR-708 and the
SMAD3 3′-UTR. As shown in Fig. 5B,
miR-708 overexpression significantly decreased luciferase activity
in the wild-type SMAD3 3′-UTR-transfected HepG2 (P=0.019) and
SMMC-7721 (P=0.022) cells compared with the cells transfected with
the mutated SMAD3 3′-UTR. These results suggest that the 3′-UTR of
SMAD3 is directly targeted by miR-708.
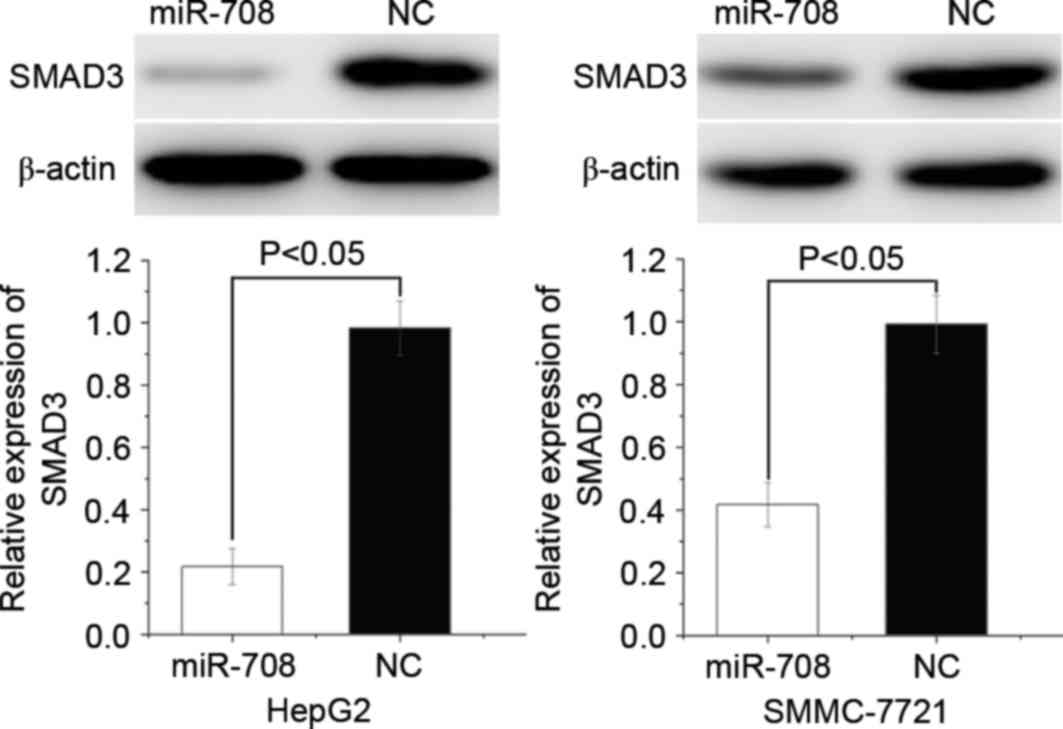
miR-708 suppresses SMAD3 protein
expression in HCC cells
To evaluate the effect of miR-708 expression on
SMAD3 protein expression, western blotting was performed in HepG2
and SMMC-7721 cells following miRNA transfection. The results
demonstrated that SMAD3 was significantly downregulated in the
miR-708-overexpressing HepG2 (P=0.013) and SMMC-7721 (P=0.017)
cells compared with the miR-NC-transfected cells (Fig. 6). These results suggest that miR-708
negatively regulates SMAD3 protein expression by directly binding
to the 3′-UTR of SMAD3 mRNA.
Discussion
Previous studies have indicated that 1/2 of miRNAs
are located in the fragile or oncogene-associated regions of
chromosomes, suggesting that abnormally expressed miRNAs are
associated with carcinogenesis and cancer progression (27). miRNAs regulate gene expression at the
post-transcriptional level via sequence-specific binding to the
3′-UTR of target mRNAs (28). A
single miRNA is able to regulate various mRNAs (29,30).
miRNAs may therefore present a novel therapeutic target in the
treatment of human cancer, including HCC (31,32). To
investigate the function of miRNAs in carcinogenesis and cancer
progression, aberrant miRNA expression must be validated, and the
biological role of miRNA in cancer initiation and progression
investigated.
miR-708 is dysregulated in multiple types of cancer,
and previous studies (22,23) reported that miR-708 was downregulated
in renal cell carcinoma and prostate cancer. Guo et al
(33) observed that miR-708
expression is low in human glioblastoma. By contrast, miR-708 was
observed to be upregulated in non-small cell lung cancer, childhood
common precursor B-cell acute lymphoblastic leukaemia and bladder
carcinoma (23,34,35). In
the present study, it was demonstrated that miR-708 expression was
decreased in HCC tissue samples and cell lines. Decreased miR-708
expression was associated with HCC tumour stage. These results
suggest that miR-708 expression is tissue specific.
miR-708 also has important roles in several types of
human cancer. In human glioblastoma, ectopic expression of miR-708
suppresses cell growth and invasion, and increases apoptosis by
negatively regulating multiple target mRNAs, including AKT
serine/threonine kinase 1, cyclin D1, matrix metalloproteinase 2,
enhancer of zeste 2 polycomb repressive complex 2 subunit, poly
(ADP-ribose) polymerase 1 and B-cell lymphoma 2 apoptosis regulator
(33). In renal cell carcinoma,
ectopic expression of miR-708 represses cell proliferation,
clonality and motility, and increases apoptosis by inhibiting zinc
finger E-box binding homeobox 2 and BMI1 proto-oncogene polycomb
ring finger (22). In prostate
cancer, downregulation of miR-708 leads to a significant increase
in tumour occurrence and development, through the direct targeting
of cluster of differentiation 44 and AKT2 (36). In addition, Li et al (37) reported that miR-708 was downregulated
in HCC and was associated with increased Edmondson-Steiner grading
and tumour node metastasis stage. In the present study, ectopic
expression of miR-708 was demonstrated to repress HCC motility.
These findings indicate that miR-708 is a tumour suppressor in
human cancer. However, miR-708 has also been demonstrated to
function as an oncogene in human cancers. For example, in non-small
lung cancer, miR-708 expression is significantly associated with an
increased risk of death following adjustment for
clinically-relevant factors, including age, gender and tumour stage
(23). Furthermore, decreased miR-708
expression has been demonstrated to decrease cell growth and
metastasis in vitro by targeting transmembrane 88 mRNA
(23). In the present study, ectopic
expression of miR-708 suppressed cell proliferation, migration and
invasion by inhibiting SMAD3 protein expression. miR-708 serves
important roles in multiple types of cancer, and may therefore be a
therapeutic target in their treatment.
The identification of cancer-specific miRNAs and
their target genes is essential in understanding the role of these
miRNAs in HCC carcinogenesis and progression, and in developing
novel targeted therapies. In the present study, SMAD3 mRNA was
identified to be a direct target of miR-708 in HCC. Bioinformatic
analysis predicted SMAD3 to be a target of miR-708 and this was
confirmed using luciferase reporter assays, which demonstrated
direct binding of miR-708 to the SMAD3 3′-UTR. Western blotting
demonstrated that miR-708 reduced SMAD3 protein expression in HCC
cell lines. These findings suggest that miR-708 acts as a tumour
suppressor in HCC by directly targeting SMAD3.
The transforming growth factor (TGF)-β signalling
pathway has an important function in numerous cellular processes,
including differentiation, growth, evasion of immunosurveillance,
metastasis and neoplasia (38,39). The
TGF-β signalling pathway is mediated by a type I receptor, a type
II receptor and SMAD proteins (40).
SMAD3 is the central mediator of the TGF-β signalling pathway
(40) and functions as an oncogene in
numerous types of cancer. In lung carcinoma, decreased SMAD3
expression significantly decreases cell migration and invasion
(25). SMAD3 also serves an important
role in epithelial-to-mesenchymal transition, which is essential in
metastasis (26). In HCC, SMAD3 is
upregulated and significantly associated with poor prognosis
(41). Therefore, investigation into
novel SMAD3-targeted HCC therapies is warranted.
SMAD3 is regulated by multiple miRNAs in numerous
types of cancer. In colorectal cancer, miR-140 decreases cell
migration and invasion through the regulation of SMAD3 (42). Liu et al (43) reported that miR-34b functions as a
tumour suppressor in pancreatic cancer by repressing SMAD3 protein
expression. In lung adenocarcinoma, ectopic expression of miR-136
inhibits metastasis by targeting SMAD3 (44). Furthermore, in nasopharyngeal cancer,
miR-145 suppresses metastasis through the inhibition of SMAD3
protein expression (45). In the
present study, miR-708 negatively regulated SMAD3 protein
expression, leading to a subsequent reduction in HCC cell
proliferation, migration and invasion. miR-708 may therefore be a
novel target of HCC treatment.
In conclusion, the present study demonstrates that
miR-708 is significantly downregulated in HCC and associated with
increased tumour stage. miR-708 decreases cell proliferation,
migration and invasion by directly targeting SMAD3 mRNA in HCC.
Identification of miR-708 targets may provide an in-depth
understanding of the potential underlying mechanisms of
carcinogenesis and progression in HCC. miR-708 may be a novel
target for future HCC therapy.
References
|
1
|
Liu H, Li W, Chen C, Pei Y and Long X:
MiR-335 acts as a potential tumor suppressor miRNA via
downregulating ROCK1 expression in hepatocellular carcinoma. Tumour
Biol. 36:6313–6319. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2015. CA Cancer J Clin. 65:5–29. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Huitzil-Melendez FD, Capanu M, O'Reilly
EM, Duffy A, Gansukh B, Saltz LL and Abou-Alfa GK: Advanced
hepatocellular carcinoma: Which staging systems best predict
prognosis? J Clin Oncol. 28:2889–2895. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Vertemati M, Moscheni C, Petrella D,
Lamperti L, Cossa M, Gambacorta M, Goffredi M and Vizzotto L:
Morphometric analysis of hepatocellular nodular lesions in HCV
cirrhosis. Pathol Res Pract. 208:240–244. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Liang T, Chen EQ and Tang H: Hepatitis B
virus gene mutations and hepatocarcinogenesis. Asian Pac J Cancer
Prev. 14:4509–4513. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Gao J, Xie L, Yang WS, Zhang W, Gao S,
Wang J and Xiang YB: Risk factors of hepatocellular
carcinoma-current status and perspectives. Asian Pac J Cancer Prev.
13:743–752. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Yang LY, Fang F, Ou DP, Wu W, Zeng ZJ and
Wu F: Solitary large hepatocellular carcinoma: A specific subtype
of hepatocellular carcinoma with good outcome after hepatic
resection. Ann Surg. 249:118–123. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
El-Serag HB and Rudolph KL: Hepatocellular
carcinoma: Epidemiology and molecular carcinogenesis.
Gastroenterology. 132:2557–2576. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Furuta M, Kozaki KI, Tanaka S, Arii S,
Imoto I and Inazawa J: miR-124 and miR-203 are epigenetically
silenced tumor-suppressive microRNAs in hepatocellular carcinoma.
Carcinogenesis. 31:766–776. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Iwakawa HO and Tomari Y: Molecular
insights into microRNA-mediated translational repression in plants.
Mol Cell. 52:591–601. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Engels BM and Hutvagner G: Principles and
effects of microRNA-mediated post-transcriptional gene regulation.
Oncogene. 25:6163–6169. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Niyazi M, Zehentmayr F, Niemöller OM,
Eigenbrod S, Kretzschmar H, Schulze-Osthoff K, Tonn JC, Atkinson M,
Mörtl S and Belka C: MiRNA expression patterns predict survival in
glioblastoma. Radiat Oncol. 6:1532011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Chen G, Lu L, Liu C, Shan L and Yuan D:
MicroRNA-377 suppresses cell proliferation and invasion by
inhibiting TIAM1 expression in hepatocellular carcinoma. PLoS One.
10:e01177142015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Tahara H, Kay MA, Yasui W and Tahara E:
MicroRNAs in Cancer: The 22nd Hiroshima Cancer Seminar/the 4th
Japanese Association for RNA Interference Joint International
Symposium, 30 August 2012, Grand Prince Hotel Hiroshima. Jpn J Clin
Oncol. 43:579–582. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yates LA, Norbury CJ and Gilbert RJ: The
long and short of microRNA. Cell. 153:516–519. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhang W, Liu K, Liu S, Ji B, Wang Y and
Liu Y: MicroRNA-133a functions as a tumor suppressor by targeting
IGF-1R in hepatocellular carcinoma. Tumour Biol. 36:9779–9788.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Chen X, Bo L, Zhao X and Chen Q:
MicroRNA-133a inhibits cell proliferation, colony formation
ability, migration and invasion by targeting matrix
metallopeptidase 9 in hepatocellular carcinoma. Mol Med Rep.
11:3900–3907. 2015.PubMed/NCBI
|
|
19
|
Li D, Liu X, Lin L, Hou J, Li N, Wang C,
Wang P, Zhang Q, Zhang P, Zhou W, et al: MicroRNA-99a inhibits
hepatocellular carcinoma growth and correlates with prognosis of
patients with hepatocellular carcinoma. J Biol Chem.
286:36677–36685. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Morishita A and Masaki T: miRNA in
hepatocellular carcinoma. Hepatol Res. 45:128–141. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Imbeaud S, Ladeiro Y and Zucman-Rossi J:
Identification of novel oncogenes and tumor suppressors in
hepatocellular carcinoma. Semin Liver Dis. 30:75–86. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Saini S, Yamamura S, Majid S, Shahryari V,
Hirata H, Tanaka Y and Dahiya R: MicroRNA-708 induces apoptosis and
suppresses tumorigenicity in renal cancer cells. Cancer Res.
71:6208–6219. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Jang JS, Jeon HS, Sun Z, Aubry MC, Tang H,
Park CH, Rakhshan F, Schultz DA, Kolbert CP and Lupu R: Increased
miR-708 expression in NSCLC and its association with poor survival
in lung adenocarcinoma from never smokers. Clin Cancer Res.
18:3658–3667. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Tian F, Byfield S DaCosta, Parks WT, Yoo
S, Felici A, Tang B, Piek E, Wakefield LM and Roberts AB: Reduction
in Smad2/3 signaling enhances tumorigenesis but suppresses
metastasis of breast cancer cell lines. Cancer Res. 63:8284–8292.
2003.PubMed/NCBI
|
|
26
|
Zavadil J and Böttinger EP: TGF-beta and
epithelial-to-mesenchymal transitions. Oncogene. 24:5764–5774.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Calin GA, Sevignani C, Dumitru CD, Hyslop
T, Noch E, Yendamuri S, Shimizu M, Rattan S, Bullrich F, Negrini M
and Croce CM: Human microRNA genes are frequently located at
fragile sites and genomic regions involved in cancers. Proc Natl
Acad Sci USA. 101:pp. 2999–3004. 2004; View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Baek D, Villén J, Shin C, Camargo FD, Gygi
SP and Bartel DP: The impact of microRNAs on protein output.
Nature. 455:64–71. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Selbach M, Schwanhäusser B, Thierfelder N,
Fang Z, Khanin R and Rajewsky N: Widespread changes in protein
synthesis induced by microRNAs. Nature. 455:58–63. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wang Y, Lu Z, Li Y, Ji D, Zhang P, Liu Q
and Yao Y: miR-143 inhibits proliferation and invasion of
hepatocellular carcinoma cells via down-regulation of TLR2
expression. Xi Bao Yu Fen Zi Mian Yi Xue Za Zhi. 30:1076–1079.
2014.(In Chinese). PubMed/NCBI
|
|
32
|
Yang XW, Zhang LJ, Huang XH, Chen LZ, Su
Q, Zeng WT, Li W and Wang Q: miR-145 suppresses cell invasion in
hepatocellular carcinoma cells: miR-145 targets ADAM17. Hepatol
Res. 44:551–559. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Guo P, Lan J, Ge J, Nie Q, Mao Q and Qiu
Y: miR-708 acts as a tumor suppressor in human glioblastoma cells.
Oncol Rep. 30:870–876. 2013.PubMed/NCBI
|
|
34
|
Song T, Zhang X, Zhang L, Dong J, Cai W,
Gao J and Hong B: miR-708 promotes the development of bladder
carcinoma via direct repression of Caspase-2. J Cancer Res Clin
Oncol. 139:1189–1198. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Li X, Li D, Zhuang Y, Shi Q, Wei W and Ju
X: Overexpression of miR-708 and its targets in the childhood
common precursor B-cell ALL. Pediatr Blood Cancer. 60:2060–2067.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Saini S, Majid S, Shahryari V, Arora S,
Yamamura S, Chang I, Zaman MS, Deng G, Tanaka Y and Dahiya R:
miRNA-708 control of CD44(+) prostate cancer-initiating cells.
Cancer Res. 72:3618–3630. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Li G, Yang F, Xu H, Yue Z, Fang X and Liu
J: MicroRNA-708 is downregulated in hepatocellular carcinoma and
suppresses tumor invasion and migration. Biomed Pharmacother.
73:154–159. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Kaminska B, Wesolowska A and Danilkiewicz
M: TGF beta signalling and its role in tumour pathogenesis. Acta
Biochim Pol. 52:329–337. 2005.PubMed/NCBI
|
|
39
|
Massagué J: TGFbeta in Cancer. Cell.
134:215–230. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Yang YA, Zhang GM, Feigenbaum L and Zhang
YE: Smad3 reduces susceptibility to hepatocarcinoma by sensitizing
hepatocytes to apoptosis through downregulation of Bcl-2. Cancer
Cell. 9:445–457. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Kim SH, Ahn S and Park CK: Smad3 and its
phosphoisoforms are prognostic predictors of hepatocellular
carcinoma after curative hepatectomy. Hepatobiliary Pancreat Dis
Int. 11:51–59. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Zhao W, Zou J, Wang B, Fan P, Mao J, Li J,
Liu H, Xiao J, Ma W, Wang M, et al: microRNA-140 suppresses the
migration and invasion of colorectal cancer cells through targeting
Smad3. Zhonghua Zhong Liu Za Zhi. 36:739–745. 2014.(In Chinese).
PubMed/NCBI
|
|
43
|
Liu C, Cheng H, Shi S, Cui X, Yang J, Chen
L, Cen P, Cai X, Lu Y, Wu C, et al: MicroRNA-34b inhibits
pancreatic cancer metastasis through repressing Smad3. Curr Mol
Med. 13:467–478. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Yang Y, Liu L, Cai J, Wu J, Guan H, Zhu X,
Yuan J, Chen S and Li M: Targeting Smad2 and Smad3 by miR-136
suppresses metastasis-associated traits of lung adenocarcinoma
cells. Oncol Res. 21:345–352. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Huang H, Sun P, Lei Z, Li M, Wang Y, Zhang
HT and Liu J: miR-145 inhibits invasion and metastasis by directly
targeting Smad3 in nasopharyngeal cancer. Tumour Biol.
36:4123–4131. 2015. View Article : Google Scholar : PubMed/NCBI
|