Introduction
As one of the most common cancer types in the world,
hepatocellular carcinoma (HCC) has an extremely high morbidity and
mortality rate, particularly in Asia and Africa (1). Overall, 50 to 55% of HCC cases are
attributable to persistent hepatitis B virus (HBV) infections,
which may result in end-stage liver disease, including liver
cirrhosis and HCC (2). As the
smallest open reading frame of the HBV genome, HBX encodes the
hepatitis B virus X (HBx) protein which has been implicated in
HBV-associated HCC pathogenesis, acting as a weak oncogene or a
cofactor in hepatocarcinogenesis (3–5). However,
the molecular mechanisms underlying HBx protein-mediated
tumorigenesis are not entirely clear. Previous studies have
demonstrated that genetic alterations alone do not account for the
complexity of HBx-induced hepatocarcinogenesis, but that epigenetic
changes, including DNA methylation (6), histone modifications (7) and non-coding RNA expression (6,8), are also
involved in this process.
Long non-coding RNAs (lncRNAs) are a type of
non-coding RNAs which are longer than 200 nucleotide transcripts
and have little or no protein-coding capacity (9,10).
Previous studies have demonstrated that lncRNAs are involved in
diverse biological functions and pathological processes (10,11), and
that altered lncRNA levels may result in aberrant gene expression
through a variety of mechanisms, including transcription,
post-transcriptional processing (12), chromatin modification, genomic
imprinting and the regulation of protein function (13). Increasing evidence demonstrates that
altered expression levels of lncRNAs contribute to a wide range of
cancer types, including breast, lung, prostate and liver cancer
(14–17). Therefore, lncRNAs may potentially be
used as diagnostic markers or therapeutic targets for cancer in the
clinic.
Using lncRNA microarrays and gene sequencing
technology, a large number of lncRNAs have been observed to be
aberrantly expressed in HCC tissues and involved in
hepatocarcinogenesis. These include highly upregulated in liver
cancer (HULC), high expression in HCC (HEIH), activated by TGF-β
(ATB) and HOX transcript antisense RNA (HOTAIR), which serve a role
in diverse biological processes including cell proliferation,
apoptosis and metastasis (17–20).
Several lncRNAs have been identified to be associated with the HBx
protein (17,21). Huang et al (22) examined the lncRNA expression profiles
in the livers of HBx transgenic and wild-type mice, and observed
that certain lncRNAs are dysregulated and associated with HBx in
HBx transgenic mice. These authors further investigated the
biological function of the lncRNA Dreh, which may be downregulated
by HBx protein, in mice. It was observed to inhibit HCC growth and
metastasis, acting as a tumor suppressor in the development of
HBV-HCC. The same authors also identified a human ortholog of Dreh,
which was termed DREH, and observed that its expression level was
frequently downregulated in HBV-associated HCC tissues. This
decrement was significantly correlated with poor survival in HCC
patients. However, the specific role of lncRNA DREH in HCC remains
largely unknown.
In the present study, the expression levels of
lncRNA DREH in 30 pairs of human HBV-positive HCC tissues and 30
pairs of HBV-negative HCC tissues and their pair-matched normal
liver tissues were assessed. The results revealed that the
expression level of DREH was significantly downregulated in HBV-HCC
tissues compared with their adjacent non-cancerous hepatic tissues,
and was inversely correlated with HBx mRNA expression in
HBV-associated HCCs. Further investigation of the biological
function of DREH in vivo and in vitro revealed that
inhibition of DREH promotes cell proliferation in HBx-induced
hepatocarcinogenesis. Together, these results suggest that DREH
exerts an impact as a potential tumor repressor gene and may
provide new insight into the role of HBx-associated lncRNAs in the
development of HCC.
Materials and methods
Animal and patient samples
The four-week-old male BALB/c nude mice used in this
study were purchased from the Experimental Animal Center of the
Chinese Academy of Medical Sciences (Beijing, China). All mice were
bred and maintained in a pathogen-free facility and were used in
accordance with the institutional guidelines for animal care. The
animal studies were approved by the Institutional Animal Care and
Use Committee of the Capital Medical University, Beijing,
China.
The 30 HBV-associated HCC tissues and 30
HBV-negative HCC tissues and corresponding adjacent non-cancerous
liver tissues used in this study were obtained with informed
consent from patients who underwent radical resection in the Peking
University People's Hospital (Beijing, China). Studies using human
tissues were reviewed and approved by the Committees for Ethical
Review of Research Involving Human Subjects of the Capital Medical
University. The clinicopathological characteristics of the 60
patients are summarized in Table
I.
 | Table I.Clinicopathological characteristics
of 60 HCC patients. |
Table I.
Clinicopathological characteristics
of 60 HCC patients.
| Characteristic | Number (n=60) | Percentage |
|---|
| Age (years) |
|
|
|
≤55 | 39 | 65.00 |
|
>55 | 21 | 35.00 |
| Sex |
|
|
|
Male | 51 | 85.00 |
|
Female | 9 | 15.00 |
| Tumor
differentiation |
|
|
|
I–II | 21 | 35.00 |
|
III–IV | 39 | 65.00 |
| TNM stage |
|
|
| I | 25 | 41.67 |
|
II–III | 35 | 58.33 |
| Tumor size
(cm) |
|
|
| ≤5 | 31 | 51.67 |
|
>5 | 29 | 48.33 |
| Tumor number |
|
|
|
Single | 52 | 86.67 |
|
Multiple | 8 | 13.33 |
| AFP (µg/l) |
|
|
|
≤20 | 12 | 20.00 |
|
>20 | 48 | 80.00 |
| Encapsulation |
|
|
|
Absent | 29 | 48.33 |
|
Complete | 31 | 51.67 |
| Microvascular
invasion |
|
|
|
Absent | 51 | 85.00 |
|
Present | 9 | 15.00 |
| Macrovascular
invasion |
|
|
|
Absent | 54 | 90.00 |
|
Present | 6 | 10.00 |
| Liver
cirrhosis |
|
|
|
Absent | 13 | 21.67 |
|
Present | 47 | 78.33 |
| HBs antigen |
|
|
|
Negative | 30 | 50.00 |
|
Positive | 30 | 50.00 |
| HBe antigen |
|
|
|
Negative | 40 | 66.67 |
|
Positive | 20 | 33.33 |
| ALT (U/l) |
|
|
|
≤40 | 26 | 43.33 |
|
>40 | 34 | 56.67 |
Construction of vectors
To construct HBx-expressing vectors, complementary
DNA encoding HBx was PCR-amplified and sub-cloned into the pcDNA3.1
vector (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA,
USA). All vectors were constructed according to standard methods
and verified by sequencing. The polymerase chain reaction (PCR)
primers used are presented in Table
II.
 | Table II.Sequences of primers and siRNAs used
in study. |
Table II.
Sequences of primers and siRNAs used
in study.
| Name |
| Sequences |
|---|
| qPCR primers |
|
|
| HBx | Sense |
5′-CCCTTCTTCATCTACCGTTCC-3′ |
|
| Anti-sense |
5′-CGTTGACATTGCTGCGAGT-3′ |
|
β-actin | Sense |
5′-TGTGTTGGCGTACAGGTCTTTG |
|
| Anti-sense |
5′-GGGAAATCGTGCGTGACATTAAG |
| DREH | Sense |
5′-CATTTGGCGGGACTACTTATT-3′ |
|
| Anti-sense |
5′-TTCAATCTGGCTTTGTTCGTT-3′ |
| Primers for vector
construction |
|
|
| DREH clone | Sense |
5′-GGGGTACCCCATGGCTGCTAGGGTGTG-3′ |
|
| Anti-sense |
5′-CGGGATCCCGTCAGGCAGAGGTGAAAAAG-3′ |
| siRNA
sequences |
|
|
| DREH siRNA | Sense |
5′-UCAUUUGGCGGGACUACUUTT-3′ |
|
| Anti-sense |
5′-AAGUAGUCCCGCCAAAUGATT-3′ |
| siRNA NC | Sense |
5′-UUCUCCGAACGUGUCACGUTT-3′ |
|
| Anti-sense |
5′-ACGUGACACGUUCGGAGAATT-3′ |
| HBx siRNA | Sense |
5′-CCCACCAAAUAUUGCCCAATT-3′ |
|
| Anti-sense |
5′-UUGGGCAAUAUUUGGUGGGTT-3′ |
Cell culture and transfection
The liver cell lines HepG2, HepG2.2.15, Hep3B, Huh-7
and SMMC-7721 were obtained from the American Type Culture
Collection (Manassas, VA, USA). The cells were grown in Dulbecco's
modified Eagle's medium (Gibco; Thermo Fisher Scientific, Inc.)
with 10% fetal bovine serum (Gibco) and were maintained in a
humidified 37°C incubator with an atmosphere of 5% CO2.
The different plasmids and small interfering RNA (siRNA) sequences
were transfected into cells using a Lipofectamine® 3000 kit
(Invitrogen) according to the manufacturer's protocol. The siRNAs
were synthesized by GenePharma (Shanghai, China). The siRNA
sequences are provided in Table
II.
Reverse transcription and quantitative
PCR (RT-qPCR)
Total RNA was extracted using TRIzol reagent
(Invitrogen). First-strand cDNA was generated using the Reverse
Transcription system kit (Stratagene, La Jolla, CA, USA). Random
primers (6mer; Takara Bio, Inc., Otsu, Japan) were used for RT-PCR
for lncRNAs. Real-time PCR was performed using a standard
SYBR-Green PCR kit protocol on a StepOne Plus system (Applied
Biosystems, Thermo Fisher Scientific, Inc.). β-actin was employed
as an endogenous control to normalize for the amount of total mRNA
in each sample. The qPCR reactions were performed in triplicate,
including no-template controls. The relative RNA expression was
calculated using the comparative Cq method. The primer sequences
are presented in Table II.
Cell Counting Kit-8 (CCK-8) assay
HepG2 or Huh-7 cells (2×103 cells/well)
transfected with DREH siRNA or negative control were dispensed in
100-µl aliquots into 96-well plates. At the indicated time points,
CCK-8 (Dojindo Molecular Technologies, Inc., Kumamoto, Japan) was
added to the cells for 2 h and then the optical density was read
using a microplate reader (Bio-Rad Laboratories, Inc., Hercules,
CA, USA). All of the experiments were performed in triplicate.
Colony formation assay
For colony formation assay, cells were seeded at a
density of 100 cells per well in a 12-well culture plate and
cultured for 2 weeks, then cells were washed twice with
phosphate-buffered saline (PBS), fixed with methanol, and the
colonies were stained with 1% crystal violet and counted.
In vivo assay for tumor growth
Lentivirus-based short hairpin RNA (shRNA)
constructs (GenePharma) were used to stably knock down DREH gene
expression according to the manufacturer's protocol. HepG2 cells
were stably transduced with DREH shRNA lentivirus. Cells
transfected with DREH shRNA or control shRNA (1.0×107)
were suspended in 100 µl PBS and implanted subcutaneously into the
bilateral armpit of BALB/c nude mice (five in each group). The
tumors were measured every three days after implantation, and the
volume of each tumor was calculated as: Length × width2
× 0.4. All mice were sacrificed four weeks later.
Statistical analysis
The expression of DREH in HCC patients was compared
using the paired samples t-test. The association between DREH and
HBx mRNA expression was analyzed by Pearson's correlation. The
correlations between DREH and clinicopathological characteristics
in the 60 HCC patients were analyzed by the χ2 test or
Fisher's exact probability test. Others comparisons were determined
by Student's t-test. All P-values were two-sided and obtained using
the SPSS 18.0 software package (SPSS, Inc., Chicago, IL, USA).
P<0.05 was considered to indicate a statistically significant
difference.
Results
LncRNA DREH is significantly
downregulated in HBV-associated HCC tissues
To confirm the role of lncRNA DREH in HCC, DREH
expression levels were first examined in 30 pairs of human
HBV-associated HCC tissues and 30 pairs of HBV-negative HCC tissues
and their pair-matched normal liver tissues by qPCR. The results
revealed that the expression levels of DREH were significantly
downregulated in HBV-HCC tissues in comparison with adjacent
non-cancerous hepatic tissues from the same patient (P<0.0001,
paired samples t-test); however, no significant difference was
observed in the expression levels between the HBV-negative HCC
tissues and the adjacent non-cancerous hepatic tissues. In
addition, the expression of DREH was significantly higher in
HBV-negative HCC tissues compared with HBV-positive HCC tissues
(Fig. 1A).
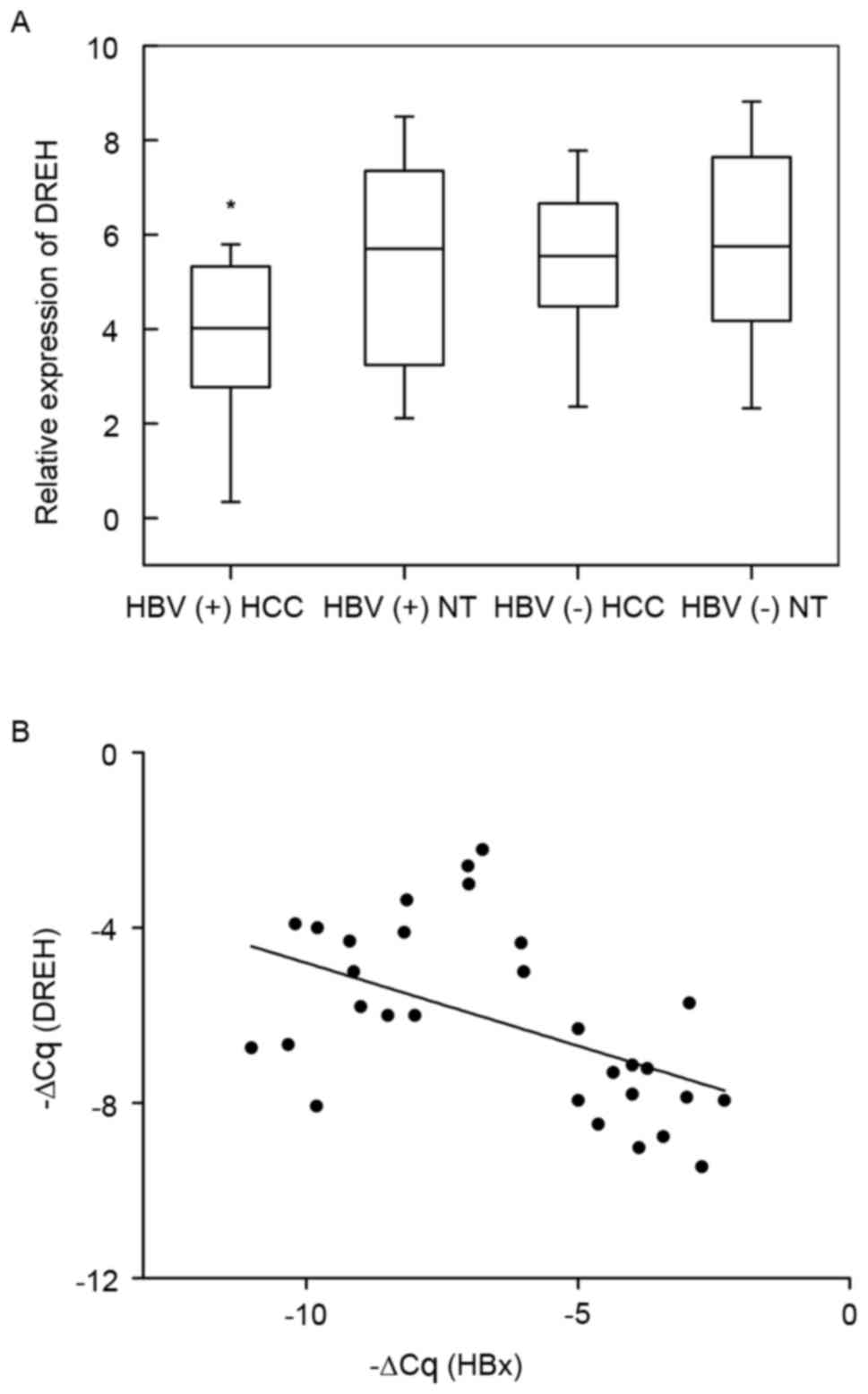 | Figure 1.LncRNA DREH is significantly
downregulated in HBV-associated HCC tissues. (A) LncRNA DREH
expression in HCC tissues vs. paired adjacent non-cancerous hepatic
tissues by qPCR (from 30 pairs of HBV-associated HCC patients and
30 pairs of HBV-negative HCC patients). Statistical differences
between HBV-HCC tissues and paired adjacent non-cancerous hepatic
tissues were analyzed with the paired samples t-test
(*P<0.0001). (B) LncRNA DREH and HBx mRNA expression levels were
inversely correlated in 30 HBV-associated HCC samples. DREH and HBx
expression levels in these samples were measured by qPCR, and
respective ΔCq values normalized to β-actin were subjected to a
Pearson correlation analysis (n=30, r=−0.531, P=0.0033, Pearson's
correlation). LncRNA, long non-coding RNA; HBV, hepatitis B virus;
HCC, hepatocellular carcinoma; qPCR, quantitative polymerase chain
reaction; HBx, hepatitis B virus X; NT, normal tissues. |
DREH expression was further compared with
clinicopathological characteristics in these 60 HCC patients, and
statistical analysis revealed that lower DREH expression levels in
HCC tissues were significantly positively correlated with tumor
size (χ2=5.406, P=0.020, Table III) and hepatitis B surface antigen
(HBsAg) (χ2=4.267, P=0.039, Table III). However, no direct correlation
was identified between the expression of lncRNA DREH and other
clinical characteristics, including age, sex, tumor
differentiation, tumor-node-metastasis stage, tumor number,
α-fetoprotein (AFP), encapsulation, microvascular invasion,
macrovascular invasion, liver cirrhosis, hepatitis B envelope
antigen and alanine aminotransferase (ALT) (Table III). These results indicate that
DREH may be involved in HCC tumor growth and potentially associated
with HBV infection.
 | Table III.Correlation between lncRNA DREH
expression and clinicopathological characteristics in 60 HCC
patients. |
Table III.
Correlation between lncRNA DREH
expression and clinicopathological characteristics in 60 HCC
patients.
|
| LncRNA DREH
expression |
|
|---|
|
|
|
|
|---|
| Characteristic | Low (n=30) | High (n=30) | P-value |
|---|
| Age (years) |
|
| 0.787 |
|
≤55 | 20 | 19 |
|
|
>55 | 10 | 11 |
|
| Sex |
|
| 0.278 |
|
Male | 24 | 27 |
|
|
Female | 6 | 3 |
|
| Tumor
differentiation |
|
| 0.787 |
|
I–II | 10 | 11 |
|
|
III–IV | 20 | 19 |
|
| TNM stage |
|
| 0.190 |
| I | 10 | 15 |
|
|
II–III | 20 | 15 |
|
| Tumor size
(cm) |
|
| 0.020a |
| ≤5 | 20 | 11 |
|
|
>5 | 10 | 19 |
|
| Tumor number |
|
| 0.254b |
|
Single | 24 | 28 |
|
|
Multiple | 6 | 2 |
|
| AFP (µg/l) |
|
| 0.333b |
|
≤20 | 4 | 8 |
|
|
>20 | 26 | 22 |
|
| Encapsulation |
|
| 0.196 |
|
Absent | 12 | 17 |
|
|
Complete | 18 | 13 |
|
| Microvascular
invasion |
|
| 1.000b |
|
Absent | 25 | 26 |
|
|
Present | 5 | 4 |
|
| Macrovascular
invasion |
|
| 0.671b |
|
Absent | 26 | 28 |
|
|
Present | 4 | 2 |
|
| Liver
cirrhosis |
|
| 0.754 |
|
Absent | 6 | 7 |
|
|
Present | 24 | 23 |
|
| HBs antigen |
|
| 0.039a |
|
Negative | 11 | 19 |
|
|
Positive | 19 | 11 |
|
| HBe antigen |
|
| 0.273 |
|
Negative | 18 | 22 |
|
|
Positive | 12 | 8 |
|
| ALT (U/l) |
|
| 0.297 |
|
≤40 | 11 | 15 |
|
|
>40 | 19 | 15 |
|
DREH and HBx mRNA levels are inversely
correlated in human HBV-associated HCC tissues
Next, it was assessed whether decreased DREH
expression was correlated with the levels of HBx expression in
human HBV-associated HCC tissues. The expression levels of HBx were
further analyzed in the aforementioned 30 HCC tissues. A
statistically significant inverse correlation was observed between
DREH and HBx mRNA (n=30, r=−0.531, P=0.0033, Pearson's correlation;
Fig. 1B). These data reveal the
potential reciprocal regulation of DREH expression induced by HBx
in human HCCs, and suggest that DREH may be involved in HCC
pathogenesis as a tumor suppressor subsequent to HBx overexpression
in chronic hepatitis B patients.
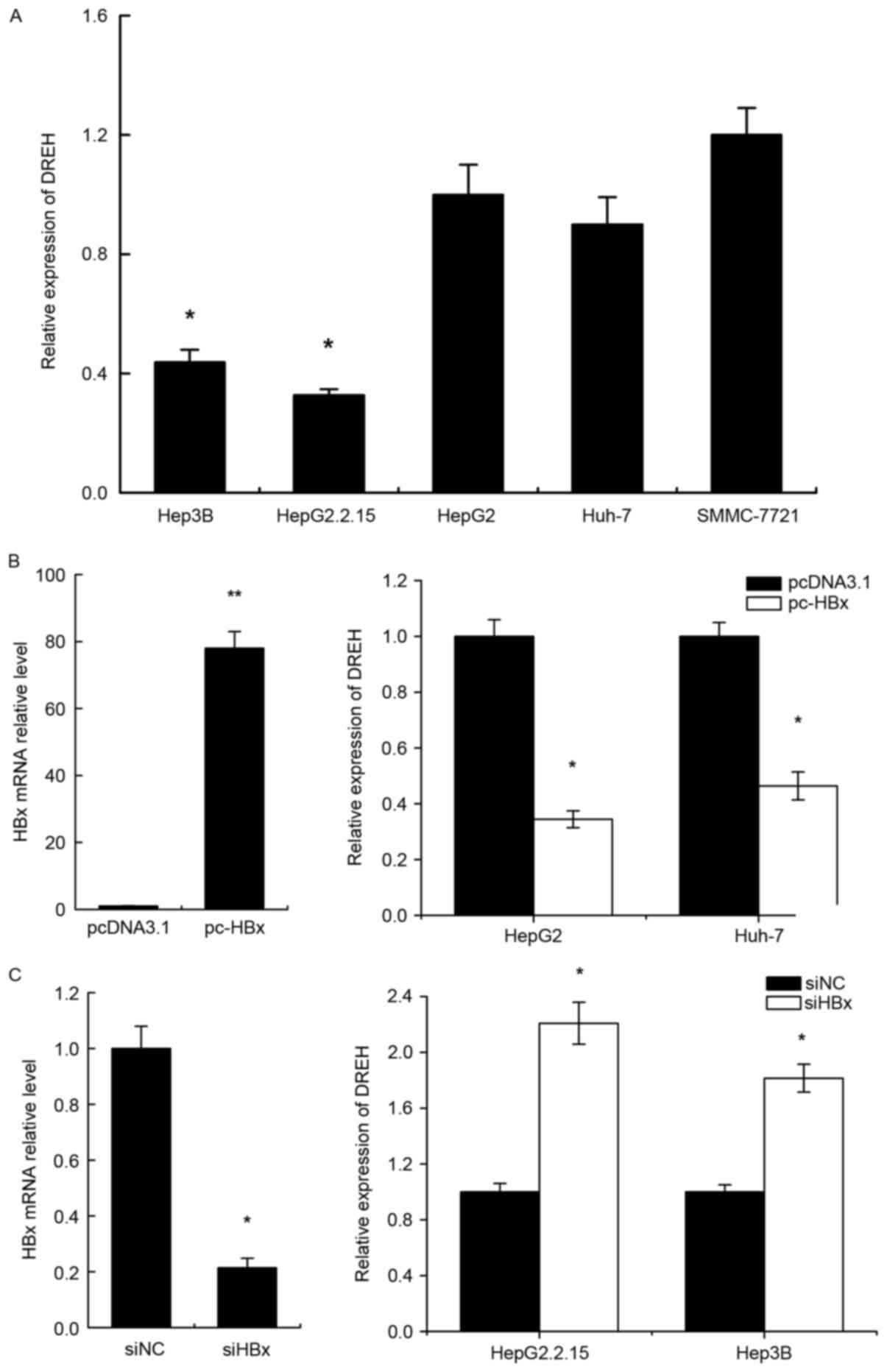
DREH is significantly downregulated in
human HCC cell lines expressing HBx
To investigate the correlation between HBx and DREH
expression, it was first determined whether DREH was differentially
expressed in human HCC cells. The expression levels of DREH were
assessed by RT-qPCR. The results revealed that the expression of
DREH was markedly lower in HepG2.2.15 (a derivative of the human
hepatoma cell line HepG2 that has been stably transformed with a
head-to-tail dimer of HBV DNA) and Hep3B (a cell line containing
the integrated hepatitis B viral genome) cell lines compared with
HepG2, Huh-7 and SMMC-7721 cells, which do not express HBx
(Fig. 2A).
Enforced HBx expression downregulates
DREH in human HCC cells
In order to verify whether this downregulation was
correlated with HBx expression, HepG2 and Huh-7 cells were
transiently transfected with HBx expression vector pc-HBx and
control vector pcDNA3.1. The levels of DREH were measured 72 h
after transient transfection. The mRNA expression of HBx following
infection is shown in the left panel of Fig. 2B. The results reveal that DREH was
downregulated in pc-HBx-transfected cells in comparison with the
pcDNA3.1 control groups (Fig.
2B).
Conversely, HBx expression was also repressed by
siRNA; the knockdown efficacy of HBx siRNA is shown in the left
panel of Fig. 2C. The inhibition of
HBx by siRNA was observed to increase DREH expression in HepG2.2.15
and Hep3B cells which express HBx (Fig.
2C).
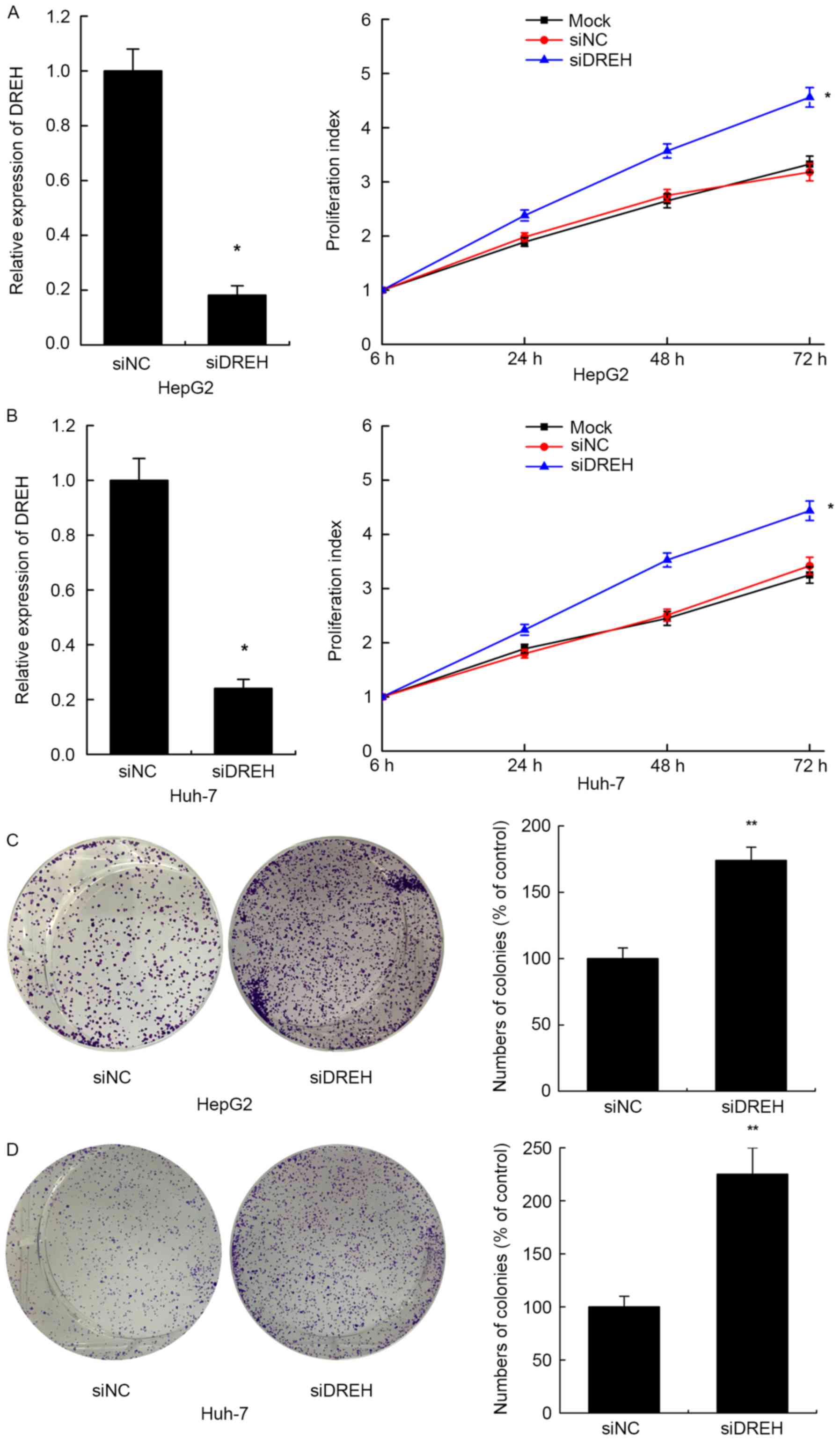
Inhibition of DREH promotes cell
proliferation of HCC cells in vitro
The frequent downregulation of lncRNA DREH by HBx
and the inverse correlation between DREH expression and tumor size
in HBV-HCC patients implies that DREH may have a role in cell
proliferation in HBV-associated hepatocarcinogenesis. To prove
this, the effects of reduced expression of DREH on cell
proliferation were investigated in two HCC cell lines. DREH
expression was repressed by RNA interference, and the relative
expression levels of DREH following infection of DREH siRNA or
control siRNA are shown in the left panel of Fig. 3A and B. Cell-Counting Kit-8 assays
demonstrated that suppression of cellular DREH enhanced the cell
proliferation index compared with the control siRNA group in HepG2
and Huh-7 cells. The negative control siRNAs did not affect the
cell proliferation index compared with the mock cells with no
treatment (Fig. 3A and B).
Further colony formation assays also revealed that
downregulation of DREH significantly enhanced the colony formation
ability in HepG2 and Huh-7 cells compared with the control cells,
consistent with the above results (Fig.
3C and D). Thus, these results suggest that DREH may serve a
key role in HBx-induced hepatocellular proliferation.
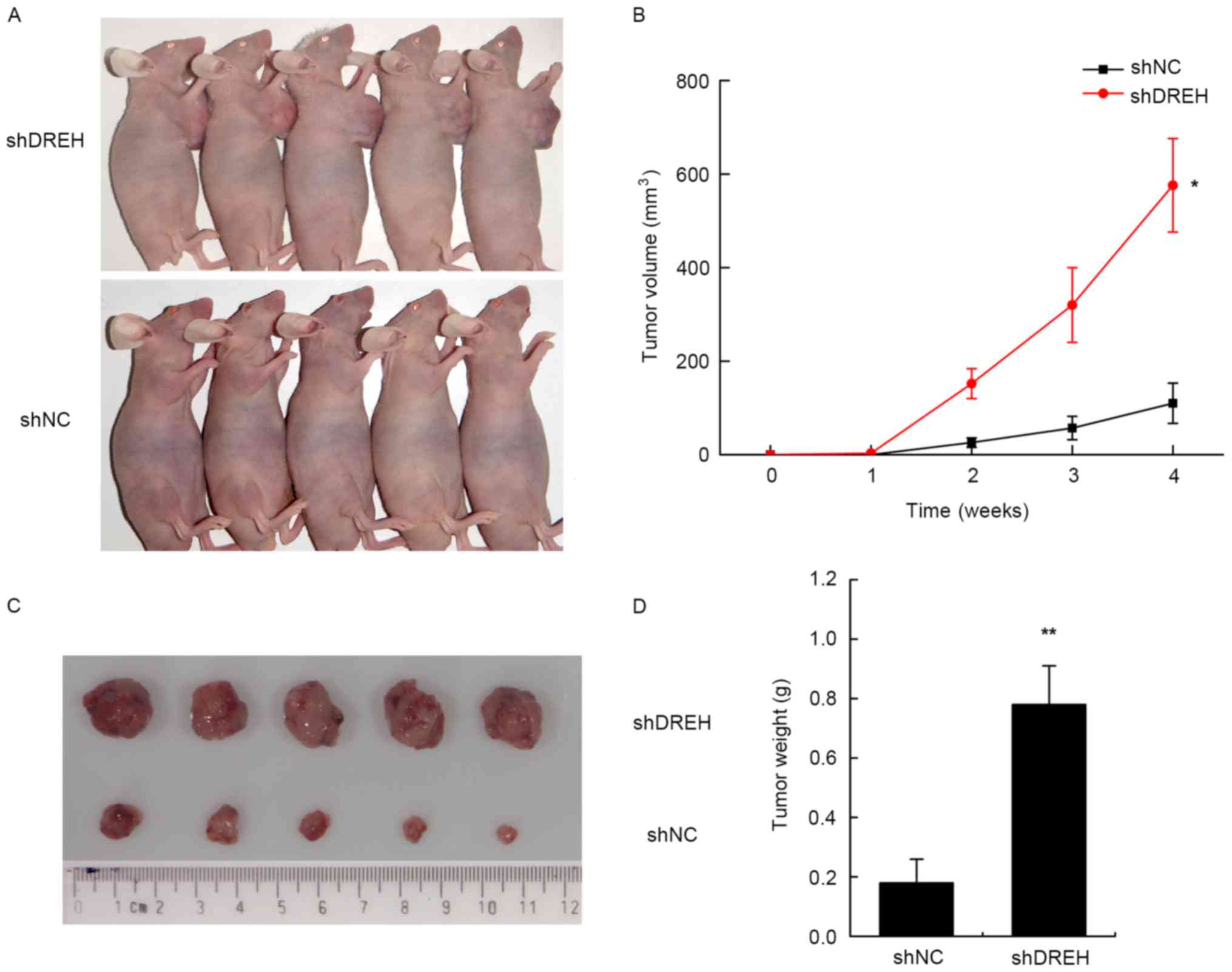
Inhibition of DREH promotes tumor
growth in vivo
To determine the effects of DREH on tumorigenesis
in vivo, DREH-downregulated or control cells (HepG2 cells
stably transfected with either shRNA-DREH or control shRNA) were
subcutaneously injected into nude mice for xenoplantation. Mice
injected with cells transfected with shRNA-DREH demonstrated
significantly increased tumor growth compared with those injected
with cells transfected with control shRNA (Fig. 4A and C).
As assessed by measurements of tumor volume, tumor
weight and tumor weight/body weight ratio, the inhibition of DREH
expression significantly promoted overall tumor growth 4 weeks
after ectopic subcutaneous implantation in nude mice (Fig. 4B and D). These results further
indicated that DREH was involved in the biological function of cell
proliferation in HBV-associated HCC.
Discussion
HCC is a leading cause of cancer-associated
mortality worldwide (23). Current
guidelines recommend different therapeutic measures for the
treatment of HCC patients with different stages, including surgery,
chemotherapy, radiation therapy, and sorafenib and transarterial
chemoembolization (24,25). Despite several recent advances and
technical refinements, the long-term survival outcome of patients
remains unsatisfactory (26).
Therefore, it is necessary to thoroughly investigate the
pathogenetic mechanism of HCC and develop new targeted treatments.
The majority of recent investigations into cancer etiology have
identified that epigenetics serves a critical role in cancer
(27,28). Alterations in epigenetic modifications
regulate all DNA-based processes, including transcription, DNA
repair and replication, and are considered to be early events in
tumorigenesis. There are also potential targets for therapeutic
intervention using epigenetic drugs (29,30).
LncRNAs are a type of epigenetic regulator and are
becoming one of the hot topics in genome research. Previous studies
have revealed various functions and molecular mechanisms of these
enigmatic molecules in biological processes of human health and
diseases (31,32). With the development of high-throughput
detection technologies including lncRNA microarray, RNA sequencing
and the recent application of next-generation sequencing, thousands
of lncRNAs have been observed to be aberrantly expressed and
associated with various cancer types (33). The HBx protein has been reported to
promote malignant transformation by epigenetic modifications and
genetic regulation during hepatocarcinogenesis (34,35). HBx
also alters the expression profiles of lncRNAs, and these
cancer-associated lncRNAs may serve key roles in gene regulation
and thus affect various aspects of cellular homeostasis (21,22).
In this study, a human lncRNA DREH was identified,
which was downregulated by HBx protein. The suppression of DREH
expression promotes the proliferation of HCC cells in vitro
and in vivo, acting as a tumor suppressor in HBx-mediated
hepatocarcinogenesis. The expression levels of DREH were examined
in 30 pairs of human HBV-positive HCC tissues and 30 pairs of
HBV-negative HCC tissues and their pair-matched normal liver
tissues. The results revealed that the expression of DREH was
frequently downregulated in HBV-associated HCC tissues compared
with their adjacent non-cancerous hepatic tissues and was inversely
correlated with HBx mRNA expression in HBV-associated HCCs.
Clinical correlation analysis demonstrated that the levels of DREH
were inversely correlated with HBsAg and tumor size in HCC
tissues.
In summary, these findings suggest that lncRNA DREH
exerts an impact as a potential tumor repressor gene in the
development of human HBV-associated HCC. The modulation of cell
proliferation by DREH may be used as a potential target for the
prevention and treatment of HBV-associated HCC.
Glossary
Abbreviations
Abbreviations:
|
HBV
|
hepatitis B virus
|
|
HBx
|
hepatitis B virus X
|
|
HCC
|
hepatocellular carcinoma
|
|
lncRNA
|
long non-coding RNA
|
|
HBsAg
|
hepatitis B surface antigen
|
|
AFP
|
α-fetoprotein
|
|
ALT
|
alanine aminotransferase
|
|
RT-qPCR
|
reverse transcription-quantitative
polymerase chain reaction
|
|
siRNA
|
small interfering RNA
|
|
CCK-8
|
Cell Counting Kit-8
|
References
|
1
|
El-Serag HB and Rudolph KL: Hepatocellular
carcinoma: Epidemiology and molecular carcinogenesis.
Gastroenterology. 132:2557–2576. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bosch FX, Ribes J, Diaz M and Cléries R:
Primary liver cancer: Worldwide incidence and trends.
Gastroenterology. 127 5 Suppl 1:1–16. 2004. View Article : Google Scholar
|
|
3
|
Koike K: Hepatitis B virus HBx gene and
hepatocarcinogenesis. Intervirology. 38:134–142. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Huang P, Zhuang B, Zhang H, Yan H, Xiao Z,
Li W, Zhang J, Tang Q, Hu K, Koeffler HP, et al: Hepatitis B virus
X protein (HBx) is responsible for resistance to targeted therapies
in hepatocellular carcinoma: Ex vivo culture evidence. Clin Cancer
Res. 21:4420–4430. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Rajput P, Shukla SK and Kumar V: The HBx
oncoprotein of hepatitis B virus potentiates cell transformation by
inducing c-Myc-dependent expression of the RNA polymerase I
transcription factor UBF. Virol J. 12:622015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Huang J, Wang Y, Guo Y and Sun S:
Down-regulated microRNA-152 induces aberrant DNA methylation in
hepatitis B virus-related hepatocellular carcinoma by targeting DNA
methyltransferase 1. Hepatology. 52:60–70. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Yuan JH, Yang F, Chen BF, Lu Z, Huo XS,
Zhou WP, Wang F and Sun SH: The histone deacetylase
4/SP1/microrna-200a regulatory network contributes to aberrant
histone acetylation in hepatocellular carcinoma. Hepatology.
54:2025–2035. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Moyo B, Nicholson SA and Arbuthnot PB: The
role of long non-coding RNAs in hepatitis B virus-related
hepatocellular carcinoma. Virus Res. 212:103–113. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Nagano T and Fraser P: No-nonsense
functions for long noncoding RNAs. Cell. 145:178–181. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wilusz JE, Sunwoo H and Spector DL: Long
noncoding RNAs: Functional surprises from the RNA world. Genes Dev.
23:1494–1504. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Mercer TR, Dinger ME and Mattick JS: Long
non-coding RNAs: Insights into functions. Nat Rev Genet.
10:155–159. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
12
|
Pandey RR, Mondal T, Mohammad F, Enroth S,
Redrup L, Komorowski J, Nagano T, Mancini-Dinardo D and Kanduri C:
Kcnq1ot1 antisense noncoding RNA mediates lineage-specific
transcriptional silencing through chromatin-level regulation. Mol
Cell. 32:232–246. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wang X, Arai S, Song X, Reichart D, Du K,
Pascual G, Tempst P, Rosenfeld MG, Glass CK and Kurokawa R: Induced
ncRNAs allosterically modify RNA-binding proteins in cis to inhibit
transcription. Nature. 454:126–130. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhao Z, Li S, Song E and Liu S: The roles
of ncRNAs and histone-modifiers in regulating breast cancer stem
cells. Protein Cell. 7:89–99. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ji P, Diederichs S, Wang W, Böing S,
Metzger R, Schneider PM, Tidow N, Brandt B, Buerger H, Bulk E, et
al: MALAT-1, a novel noncoding RNA, and thymosin beta4 predict
metastasis and survival in early-stage non-small cell lung cancer.
Oncogene. 22:8031–8041. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Fu X, Ravindranath L, Tran N, Petrovics G
and Srivastava S: Regulation of apoptosis by a prostate-specific
and prostate cancer-associated noncoding gene, PCGEM1. DNA Cell
Biol. 25:135–141. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Du Y, Kong G, You X, Zhang S, Zhang T, Gao
Y, Ye L and Zhang X: Elevation of highly up-regulated in liver
cancer (HULC) by hepatitis B virus X protein promotes hepatoma cell
proliferation via down-regulating p18. J Biol Chem.
287:26302–26311. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yang F, Zhang L, Huo XS, Yuan JH, Xu D,
Yuan SX, Zhu N, Zhou WP, Yang GS, Wang YZ, et al: Long noncoding
RNA high expression in hepatocellular carcinoma facilitates tumor
growth through enhancer of zeste homolog 2 in humans. Hepatology.
54:1679–1689. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yuan JH, Yang F, Wang F, Ma JZ, Guo YJ,
Tao QF, Liu F, Pan W, Wang TT, Zhou CC, et al: A long noncoding RNA
activated by TGF-β promotes the invasion-metastasis cascade in
hepatocellular carcinoma. Cancer Cell. 25:666–681. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ishibashi M, Kogo R, Shibata K, Sawada G,
Takahashi Y, Kurashige J, Akiyoshi S, Sasaki S, Iwaya T, Sudo T, et
al: Clinical significance of the expression of long non-coding RNA
HOTAIR in primary hepatocellular carcinoma. Oncol Rep. 29:946–950.
2013.PubMed/NCBI
|
|
21
|
Huang JL, Ren TY, Cao SW, Zheng SH, Hu XM,
Hu YW, Lin L, Chen J, Zheng L and Wang Q: HBx-related long
non-coding RNA DBH-AS1 promotes cell proliferation and survival by
activating MAPK signaling in hepatocellular carcinoma. Oncotarget.
6:33791–33804. 2015.PubMed/NCBI
|
|
22
|
Huang JF, Guo YJ, Zhao CX, Yuan SX, Wang
Y, Tang GN, Zhou WP and Sun SH: Hepatitis B virus X protein
(HBx)-related long noncoding RNA (lncRNA) down-regulated expression
by HBx (Dreh) inhibits hepatocellular carcinoma metastasis by
targeting the intermediate filament protein vimentin. Hepatology.
57:1882–1892. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Murata S, Mine T, Sugihara F, Yasui D,
Yamaguchi H, Ueda T, Onozawa S and Kumita S: Interventional
treatment for unresectable hepatocellular carcinoma. World J
Gastroenterol. 20:13453–13465. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Margini C and Dufour JF: The story of HCC
in NAFLD: From epidemiology, across pathogenesis, to prevention and
treatment. Liver Int. 36:317–324. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Greten TF, Wang XW and Korangy F: Current
concepts of immune based treatments for patients with HCC: From
basic science to novel treatment approaches. Gut. 64:842–848. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Dufour JF, Bargellini I, De Maria N, De
Simone P, Goulis I and Marinho RT: Intermediate hepatocellular
carcinoma: Current treatments and future perspectives. Ann Oncol.
24 Suppl 2:ii24–ii29. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wu Y, Sarkissyan M and Vadgama JV:
Epigenetics in breast and prostate cancer. Methods Mol Biol.
1238:425–466. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kanwal R, Gupta K and Gupta S: Cancer
epigenetics: An introduction. Methods Mol Biol. 1238:3–25. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Dawson MA and Kouzarides T: Cancer
epigenetics: From mechanism to therapy. Cell. 150:12–27. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Yang N, Ekanem NR, Sakyi CA and Ray SD:
Hepatocellular carcinoma and microRNA: New perspectives on
therapeutics and diagnostics. Adv Drug Deliv Rev. 81:62–74. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Batista PJ and Chang HY: Long noncoding
RNAs: Cellular address codes in development and disease. Cell.
152:1298–1307. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Lee JT: Epigenetic regulation by long
noncoding RNAs. Science. 338:1435–1439. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Huarte M: The emerging role of lncRNAs in
cancer. Nat Med. 21:1253–1261. 2015. View
Article : Google Scholar : PubMed/NCBI
|
|
34
|
Protzer U: Hepatitis: Epigenetic control
of HBV by HBx protein-releasing the break? Nat Rev Gastroenterol
Hepatol. 12:558–559. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Park IY, Sohn BH, Yu E, Suh DJ, Chung YH,
Lee JH, Surzycki SJ and Lee YI: Aberrant epigenetic modifications
in hepatocarcinogenesis induced by hepatitis B virus X protein.
Gastroenterology. 132:1476–1494. 2007. View Article : Google Scholar : PubMed/NCBI
|