Introduction
Osteosarcoma is the most common aggressive bone
tumor in children and adolescents (1). Although many tumors initially respond to
chemotherapy, patients with metastatic or relapsed osteosarcoma
have extremely poor survival outcomes (2). The 5-year survival rate of patients
without metastatic disease is 60–70%, while the clinical outcomes
for patients with metastatic disease are far worse, since a 5-year
survival rate of 15–30% has been reported for these patients
(3,4).
However, the mechanisms underlying the multiple oncogenic
properties of osteosarcoma are not fully understood. A
comprehensive understanding of the mechanisms is required for the
development of effective therapeutic strategies.
Previous studies have revealed that microRNAs
(miRNAs) regulate numerous cellular processes, including those
associated with cancer (5). miRNAs
are a class of small non-coding RNAs that range from 22–25
nucleotides in length (6). The
5′-region of a miRNA contains 2–8 bases, termed the seed region,
which is important for target mRNA recognition (7). miRNAs negatively regulate gene
expression by inhibiting protein translation or by causing mRNA
degradation through partial or complete base-pairing with the 3′
untranslated region (3′UTR) of the target mRNA. Dysregulation of
miRNA expression has been reported in various human cancers
(5,7).
Therefore, exploring the mechanisms underlying the carcinogenic
effects of these aberrant tumor miRNAs is critical for cancer
diagnosis and therapy.
Dysregulated miR-433 expression has been observed in
various cancers and has been significantly associated with the
clinical outcome of tumor patients. For example, the overexpression
of miR-433 in a gastric cancer cell line suppressed cell growth and
invasion by downregulating KRAS expression (8). Overexpression of miR-433 downregulated
thymidylate synthetase mRNA and protein expression and enhanced the
sensitivity of HeLa cells to 5-fluorouracil (9). In a previous study, miR-433 expression
was decreased in myeloproliferative neoplasms, and overexpression
of miR-433 was shown to inhibit hematopoietic cell growth and
differentiation by targeting guanylate binding protein 2 (GBP2)
(10). However, the role of miR-433
in osteosarcoma cells remains unknown.
In the present study, miR-433 was shown to be
upregulated in human osteosarcoma tissue compared with normal
tissue. Ectopic expression of miR-433 decreased apoptosis in
osteosarcoma cells by targeting programmed cell death 4 (PDCD4),
indicating that miR-433 may be a potential molecular target for
osteosarcoma treatment.
Materials and methods
Cell culture
Two human osteosarcoma cell lines, U2OS and
G293, were obtained from American Type Culture Collection
(Manassas, VA, USA). The normal human osteoblastic cell line hFOB
1.19 (American Type Culture Collection, Manassas, VA, USA) was used
as a control. The cells were cultured in Dulbecco's modified
Eagle's medium supplemented with 10% heat-inactivated fetal calf
serum (HyClone; GE Healthcare Life Sciences, Logan, UT, USA) at
37°C with 5% CO2 and 95% humidity.
Patients
Three paired osteosarcoma tissues and normal
adjacent tissues were obtained from each patient by biopsy prior to
any treatment. Histological diagnosis demonstrated that all tumor
samples were high-grade osteosarcoma with either stage IIA or IIB
in the Enneking surgical staging system.
RNA extraction and reverse
transcription-quantitative polymerase chain reaction (qPCR)
Total RNA was extracted from clinical specimens and
cell lines using TRIzol reagent (Thermo Fisher Scientific, Inc.,
Waltham, MA, USA), according to the manufacturer's protocol.
Reverse transcription was performed using the TaqMan MicroRNA
Reverse Transcription kit (Applied Biosystems; Thermo Fisher
Scientific, Inc.). qPCR was performed using SYBR Green Real-Time
PCR Master Mix (Roche Diagnostics, Basel, Switzerland) on the
Applied Biosystems 7500 Real-Time PCR System (Thermo Fisher
Scientific, Inc.). The cycling conditions were as follows: 94°C for
5 min, followed by 40 cycles of 94°C for 10 sec, 60°C for 45 sec
and 72°C for 30 sec. The relative expression of miR-433 was
normalized to U6 small nuclear RNA (RNU6) expression. Each
experiment was performed in triplicate. All samples were normalized
to the internal control, and fold-changes were calculated using the
2−ΔΔCq quantification method (11). The primers used for miR-433 and RNU6
(Sigma-Aldrich; Merck Millipore, Darmstadt, Germany) were as
follows: miR-433, forward 5′-GGCGGTGAATAATGAC-3′ and reverse
5′-GTGCAGGGTCCGAGGT-3′; and RNU6, forward
5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACTAAGCA-3′ and
reverse 5′-CCTGCGCAAGGATGAC-3′.
Microarray data analysis
Expression data (Gene Expression Omnibus accession
no. 39040; title: MicroRNA profiling and clinical outcomes in human
osteosarcoma) were downloaded and imported to the R environment
available at http://www.r-project.org and
processed using the Illumina-specific package lumi.
Variance-stabilizing transformation (12) and quantile normalization were applied
to analyze gene expression.
Transfections
Synthetic constructs containing miR-433 mimic,
miR-433 inhibitor, negative control or PDCD4 small interfering
(si)RNA (GenePharma Co., Ltd., Shanghai, China) were transfected
into the osteosarcoma cell lines at a final concentration of 20 nM.
All transfections were performed using Lipofectamine 2000
(Invitrogen; Thermo Fisher Scientific, Inc.), according to the
manufacturer's protocol.
Vector construction
PDCD4 was identified as a potential target for
miR-433 using TargetScan (http://www.targetscan.org/) and miRanda (http://www.microrna.org/). The fragment of the 3′UTR
of PDCD4 mRNA (GenBank accession no. NM014456.4) containing the
potential miR-433 binding site was amplified by PCR using forward
5′-TGCCATGTTTATTATCTAA-3′ and reverse 5′-AATTCGAGATCCAGTCTA-3′
primers, and cloned downstream of the luciferase reporter gene in
the pGL3-control vector (Promega Corporation, Madison, WI, USA) to
form pGL3-PDCD4-3′UTR-WT. The 3′UTR-Mut plasmid of PDCD4
(pGL3-PDCD4-3′UTR-MUT) harboring mutations in the sequence
complementary to the seed region of miR-433 was constructed using
site-specific mutagenesis (13).
Luciferase reporter assay
The dual-luciferase reporter assay was performed to
determine whether miR-433 downregulates PDCD4 by binding to the
3′UTR region. HEK293T cells (American Type Culture Collection) were
plated in a 48-well plate at 80–90% confluence. After
cotransfection with 20 nM miR-433 mimic or scramble control and 40
ng pGL3-PDCD4-3′UTR-WT or pGL3-PDCD4-3′UTR-MUT, the cells were
collected to measure Firefly luciferase activity with the
dual-luciferase assay (Promega Corporation). The assays were
performed in triplicate.
Western blotting
The cells were lysed using radioimmunoprecipitation
assay lysis buffer (Invitrogen; Thermo Fisher Scientific, Inc.)
containing 1X phenylmethylsulfonyl fluoride and 1X protease
inhibitor cocktail (Roche Diagnostics). The protein concentration
was measured using the bicinchoninic acid protein assay kit
(Pierce; Thermo Fisher Scientific, Inc.). The proteins (30 µg) were
separated by 10% SDS-PAGE and transferred onto a PVDF membrane. The
membranes were blocked with 5% nonfat milk in TBST for 1 h and
incubated overnight at 4°C with the following primary antibodies:
Rabbit monoclonal anti-β-actin (1:5,000; catalog no. 4970; Cell
Signaling Technology, Inc., Danvers, MA, USA); rabbit monoclonal
anti-PDCD4 (1:1,000; catalog no. 9535; Cell Signaling Technology,
Inc.); and rabbit monoclonal anti-PARP (1:1,000; catalog no. 9532;
Cell Signaling Technology, Inc.). Upon being washed, the membranes
were incubated with horseradish peroxidase-conjugated secondary
antibodies (1:5,000; catalog no. ab97051; Abcam, Cambridge, UK) at
room temperature for 1 h. Signals were detected with an enhanced
chemiluminescence system (Bio-Rad Laboratories, Inc., Hercules, CA,
USA).
Terminal deoxynucleotidyl transferase
dUTP nick end labeling (TUNEL) analysis
OCT-embedded cryosections (6 µm-thick) of tumors
were fixed with 4% paraformaldehyde for 30 min. Following
permeabilization with 0.1% Triton X-100 for 10 min, the tumor
sections were incubated with methanol containing 3%
H2O2, followed by dUTP nick end labeling at
37°C for 1 h using the In Situ Cell Death Detection kit
(Sigma-Aldrich; Merck Millipore).
Apoptosis analysis
Apoptosis was assessed used Annexin V/propidium
iodide (PI) staining. Adherent and floating cells were harvested by
trypsinization and washed three times with PBS. Subsequently, the
cells were stained with 5 µl Annexin V in 60 µl 1X binding buffer
(BD Biosciences, Franklin Lakes, NJ, USA) for 15 min at room
temperature in the dark. After staining, 120 µl 1X binding buffer
and 5 µl PI were added to the cells, and the cells were analyzed
using a FACSCalibur flow cytometer.
Immunofluorescence
Cells were fixed with 4% paraformaldehyde and
treated with 1:400 diluted rabbit monoclonal anti-PDCD4 antibody
(catalog no. 9535; Cell Signaling Technology, Inc.), followed by
1:400 diluted Alexa Fluor 568-conjugated anti-rabbit antibody (Cell
Signaling Technology, Inc.) and DAPI (0.5 ng/ml; Thermo Fisher
Scientific, Inc.).
Tumorigenicity assay in nude mice
A total of 10 4-to-6-week old male athymic nude
mice, which weighed 24–30 g, were purchased from Shanghai
Laboratory Animal Center (Shanghai, China). All mice were housed
and treated in accordance with the guidelines of the Shandong
University Animal Care and Use committee (Shanghai, China), and
were maintained in specific pathogen-free conditions, consisting of
a reversed 12:12-h light-dark cycle with controlled temperature (21
± 1°C) and humidity. The mice were allowed free access to food and
water. All experiments involving animals were undertaken in
accordance with the Health Guide for the Care and Use of Laboratory
Animals at Shandong University. U2OS cells (1×106 cells) stably
expressing miR-433 or the control vector were inoculated
bilaterally into the armpit region of 4-week-old immunodeficient
nude mice. The tumor weights were measured 30 days following
inoculation. All mice were euthanized by 30% CO2
inhalation for 7 min and the tumor tissues were dissected.
Statistical analysis
Data are presented as the mean ± standard deviation.
Statistical analyses were performed using SPSS 17.0 software.
Statistical significance was determined using the two-tailed
unpaired Student's t-test, unless otherwise noted. P<0.05 was
considered to indicate a statistically significant difference.
Results
miR-433 is overexpressed in
osteosarcoma
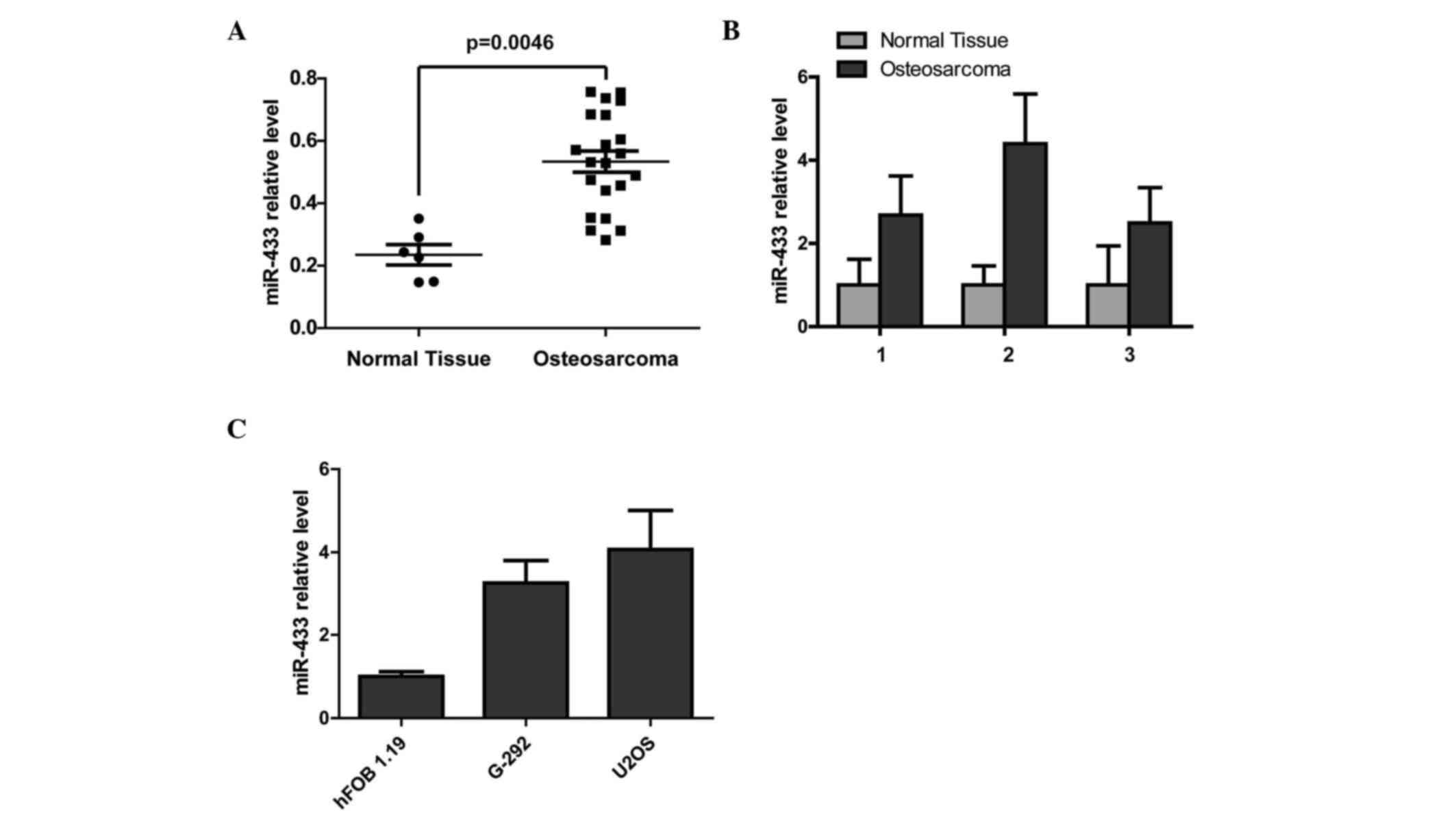
In order to detect the expression level of miR-433
in human osteosarcoma, the miRNA expression profile array (Gene
Expression Omnibus accession no. 39040; title: MicroRNA profiling
and clinical outcomes in human osteosarcoma) was analyzed and
showed that miR-433 was significantly upregulated in osteosarcoma
tissues compared with normal tissues (P=0.0046; Fig. 1A). To further confirm the upregulation
of miR-433 in osteosarcoma, RT-qPCR analysis of three matched
clinical sample pairs was performed. miR-433 expression was
markedly higher in clinical osteosarcoma tissues compared with
adjacent normal tissues (Fig. 1B). In
addition, RT-qPCR was used to show that the miR-433 expression
level in the normal human osteoblastic cell line hFOB 1.19 was
lower compared with the human osteosarcoma cell lines (Fig. 1C).
miR-433 inhibits the apoptosis of
human osteosarcoma cells in vitro
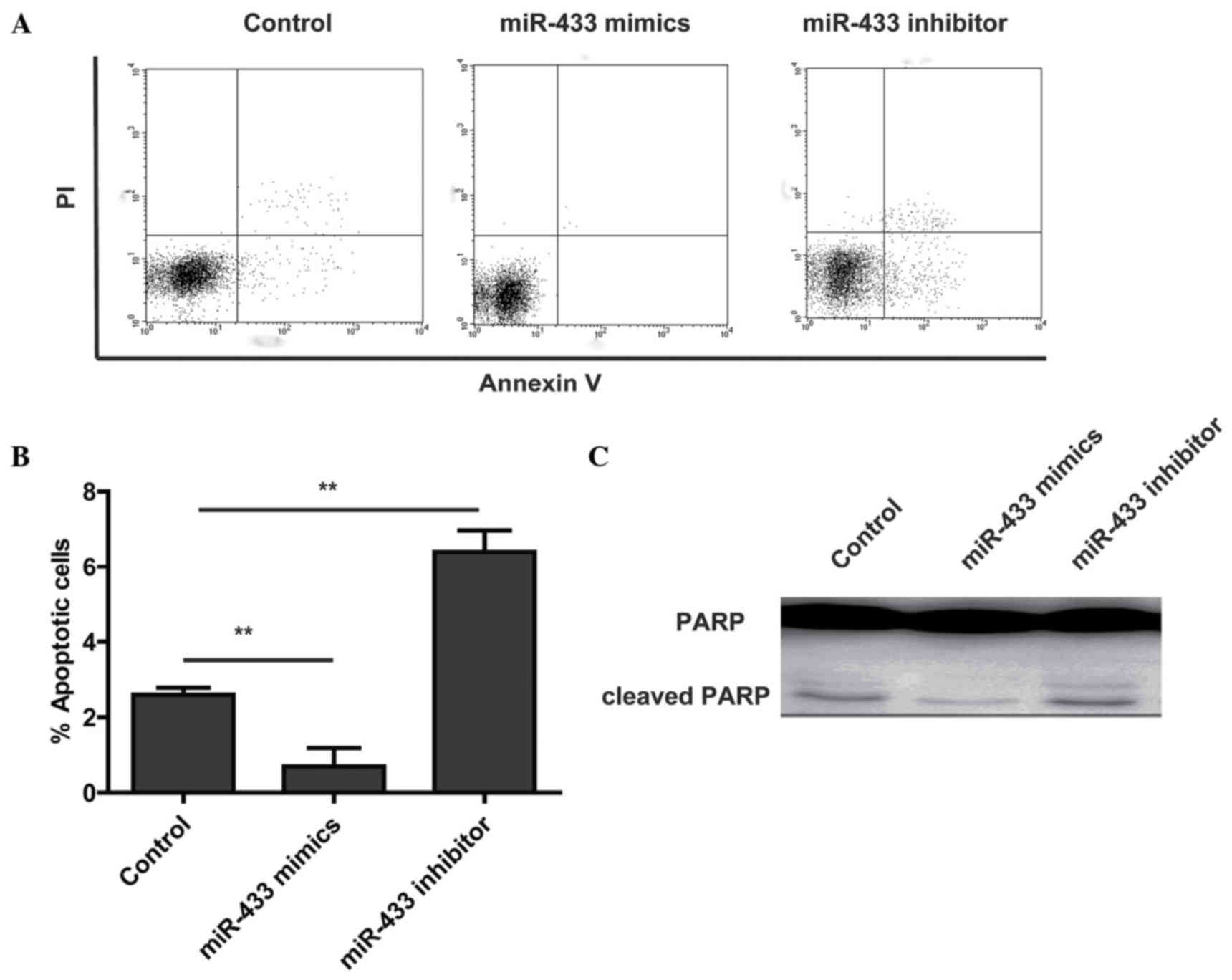
To assess the effects of miR-433 expression on
osteosarcoma cell survival, the osteosarcoma cell line U2OS was
used. Cells were transfected with miR-433 mimic, miR-433 inhibitor
or scrambled control and were stained with Annexin V, a well known
marker of early apoptosis. Flow cytometry showed that a significant
decrease in Annexin V binding was detected at 48 h following
transfection with miR-433 mimic compared with the control (Fig. 2A and B). As expected, the miR-433
inhibitor increased the apoptosis of U2OS cells compared with the
control (Fig. 2A and B). To further
confirm the effects of miR-433 on apoptosis in osteosarcoma cells,
western blot analysis to assess the levels of cleaved
poly(ADP-ribose) polymerase (PARP) was performed. The result showed
that miR-433 mimics decreased the levels of cleaved PARP (Fig. 2C). These results suggest that
overexpression of miR-433 protects the tumor cells from
apoptosis.
miR-433 suppresses the expression of
PDCD4
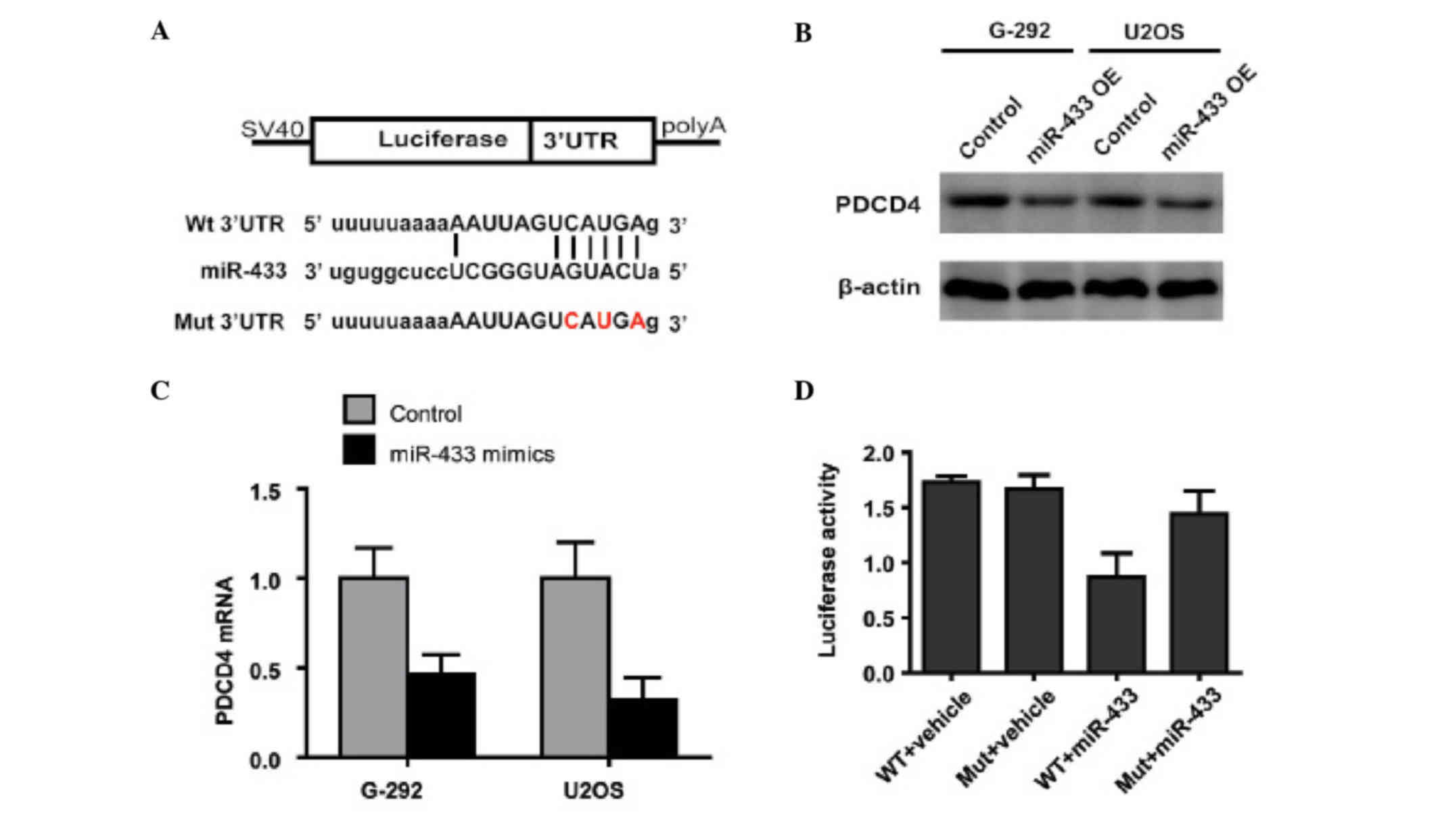
To further examine the mechanism by which miR-433
inhibits apoptosis in osteosarcoma cells, the present study
analyzed the downstream targets of miR-433 using TargetScan and
miRanda to predict potential direct targets (14,15), in
particular those involved in apoptosis. PDCD4, a tumor suppressor
involved in apoptosis (16), had
potential miR-433-binding regions in the 3′UTR of its mRNA
(Fig. 3A). RT-qPCR and western
blotting confirmed that miR-433 downregulated PDCD4 in osteosarcoma
cells at both the protein and mRNA expression levels (Fig. 3B and C), which suggested that miR-433
regulates the expression of PDCD4 by promoting the degradation of
its mRNA. To explore whether PDCD4 is indeed the target of miR-433,
the luciferase reporter assay using vectors encoding wild-type or
mutant PDCD4 3′UTR downstream of the luciferase gene was performed.
Luciferase activity was markedly decreased in
miR-433-overexpressing cells co-transfected with
pGL3-PDCD4-3′UTR-WT, but not in cells co-transfected with
pGL3-PDCD4-3′UTR-MUT (Fig. 3D).
miR-433 decreases tumor apoptosis in
vivo
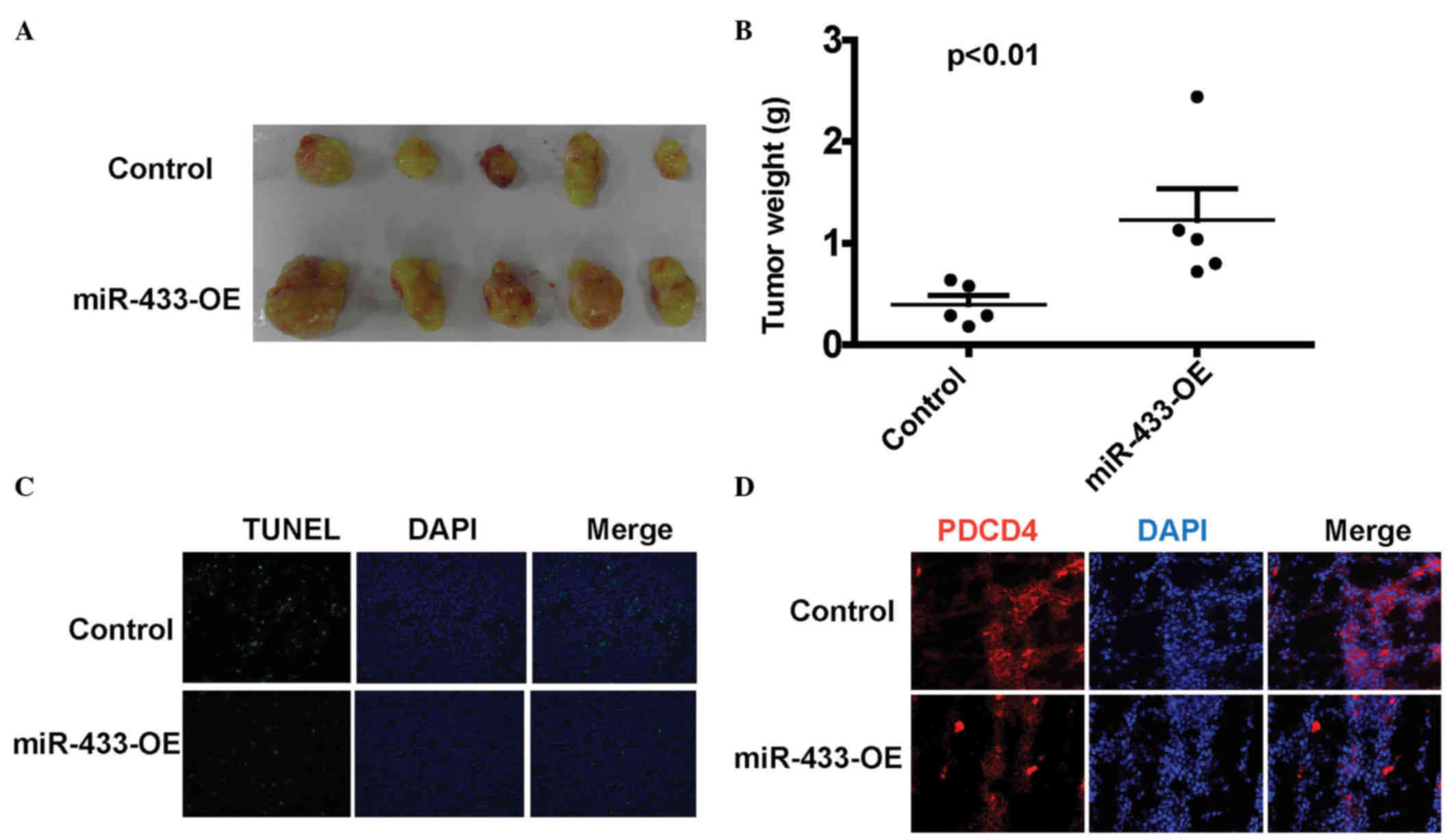
To examine the impact of miR-433 on tumor apoptosis,
Balb/c nude mice were used to establish a xenograft tumor model.
U2OS cells stably overexpressing miR-433 or vehicle were
subcutaneously injected into the bilateral armpits of nude mice.
Tumor weight was measured after sacrifice on day 30 post-injection.
As shown in Fig. 4A and B, the tumor
weight of the miR-433 overexpression group was significantly
increased compared with the control group (P=0.0073). In
situ TUNEL analysis showed that tumors overexpressing miR-433
had less TUNEL-positive cells than control group tumors (Fig. 4C). Immunofluorescence analysis of the
xenograft tumor showed that tumors overexpressing miR-433 had
decreased PDCD4 expression (Fig. 4D),
which was consistent with the in vitro data, suggesting that
miR-433 regulates apoptosis by targeting PDCD4.
Discussion
Osteosarcoma is the most common primary malignant
tumor of the bone; it has an annual worldwide incidence of 1–3
cases per 1 million (17).
Osteosarcoma occurs most frequently in children and adolescents,
with a second peak in incidence in those aged >50 years
(18). Despite significant
improvements in the diagnosis and treatment of osteosarcoma,
overall survival has remained relatively constant for >20 years
(2). Several dysregulated miRNAs,
including downregulated miR-34 and miR-143, and upregulated miR-140
and miR-21, have been associated with osteosarcoma development
(19–21). However, the identification of novel
and important dysregulated miRNAs, as well as the elucidation of
their roles in osteosarcoma carcinogenesis and progression, remains
an ongoing process.
Increasingly, studies have shown that various miRNAs
involved in apoptosis are also associated with chemotherapy
resistance in osteosarcoma (22–25). Some
miRNAs function as oncogenes, and have been reported to be
upregulated in osteosarcoma, while others serve as tumor
suppressors, and are often downregulated in osteosarcoma (26). Therefore, a potential therapeutic
strategy in osteosarcoma would be to knockdown the expression of
oncogenic miRNAs using miRNA inhibitors or to overexpress the tumor
suppressor miRNAs using miRNA mimics.
Previous reports have shown that miR-433 is
aberrantly expressed in various cancers. For example, miR-433 was
decreased in myeloproliferative neoplasms, and overexpression of
miR-433 suppressed hematopoietic cell growth and differentiation by
targeting GBP2 (10). The present
study demonstrated that miR-433 was upregulated in osteosarcoma
tissues compared with the adjacent normal tissues, and inhibited
osteosarcoma apoptosis. The present data, along with an analysis of
a published dataset (27), showed
that miR-433 was overexpressed in osteosarcoma tissues and cell
lines. In combination with previous reports revealing the roles of
miR-433 in various other types of cancer, including ovarian and
gastric cancer (28,29), the present study further confirmed
that miR-433 functions as an oncomiR in osteosarcoma. Furthermore,
overexpression of miR-433 was shown to decrease the apoptosis of
osteosarcoma cells by targeting PDCD4 in vitro. In addition,
in the xenograft tumor model, miR-433-overexpressing U2OS cells
were more resistant to apoptosis.
In conclusion, the present study is the first to
demonstrate that miR-433 is significantly overexpressed in
osteosarcoma tissues and cell lines. Furthermore, miR-433 was shown
to play a crucial role in apoptosis by targeting PDCD4 in
osteosarcoma. These results suggest that miR-433 may be a molecular
target for osteosarcoma therapy.
References
|
1
|
Ferguson WS and Goorin AM: Current
treatment of osteosarcoma. Cancer Invest. 19:292–315. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chou AJ and Gorlick R: Chemotherapy
resistance in osteosarcoma: Current challenges and future
directions. Expert Rev Anticancer Ther. 6:1075–1085. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Luetke A, Meyers PA, Lewis I and Juergens
H: Osteosarcoma treatment-where do we stand? A state of the art
review. Cancer Treat Rev. 40:523–532. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Aljubran AH, Griffin A, Pintilie M and
Blackstein M: Osteosarcoma in adolescents and adults: Survival
analysis with and without lung metastases. Ann Oncol. 20:1136–1141.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ghosh T, Aprea J, Nardelli J, Engel H,
Selinger C, Mombereau C, Lemonnier T, Moutkine I, Schwendimann L,
Dori M, et al: MicroRNAs establish robustness and adaptability of a
critical gene network to regulate progenitor fate decisions during
cortical neurogenesis. Cell Rep. 7:1779–1788. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Mitra R, Sun J and Zhao Z: microRNA
regulation in cancer: One arm or two arms? Int J Cancer.
137:1516–1518. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Treiber T, Treiber N and Meister G:
Regulation of microRNA biogenesis and function. Thromb Haemost.
107:605–610. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Guo LH, Li H, Wang F, Yu J and He JS: The
tumor suppressor roles of miR-433 and miR-127 in gastric cancer.
Int J Mol Sci. 14:14171–14184. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Gotanda K, Hirota T, Matsumoto N and Ieiri
I: MicroRNA-433 negatively regulates the expression of thymidylate
synthase (TYMS) responsible for 5-fluorouracil sensitivity in HeLa
cells. BMC Cancer. 13:3692013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lin X, Rice KL, Buzzai M, Hexner E, Costa
FF, Kilpivaara O, Mullally A, Soares MB, Ebert BL, Levine R and
Licht JD: miR-433 is aberrantly expressed in myeloproliferative
neoplasms and suppresses hematopoietic cell growth and
differentiation. Leukemia. 27:344–352. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Pfaffl MW: A new mathematical model for
relative quantification in real-time RT-PCR. Nucleic Acids Res.
29:e452001. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lin SM, Du P, Huber W and Kibbe WA:
Model-based variance-stabilizing transformation for Illumina
microarray data. Nucleic Acids Res. 36:e112008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Luo F, Du X, Weng T, Wen X, Huang J and
Chen L: Efficient multi-site-directed mutagenesis directly from
genomic template. J Biosci. 37:965–969. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Lewis BP, Burge CB and Bartel DP:
Conserved seed pairing, often flanked by adenosines, indicates that
thousands of human genes are microRNA targets. Cell. 120:15–20.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Betel D, Wilson M, Gabow A, Marks DS and
Sander C: The microRNA.org resource: targets and expression.
Nucleic Acids Res. 36:D149–D153. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lankat-Buttgereit B and Göke R: The tumour
suppressor Pdcd4: Recent advances in the elucidation of function
and regulation. Biol Cell. 101:309–317. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Chen X, Bahrami A, Pappo A, Easton J,
Dalton J, Hedlund E, Ellison D, Shurtleff S, Wu G, Wei L, et al:
Recurrent somatic structural variations contribute to tumorigenesis
in pediatric osteosarcoma. Cell Rep. 7:104–112. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Duong LM and Richardson LC: Descriptive
epidemiology of malignant primary osteosarcoma using
population-based registries, United States, 1999–2008. J Registry
Manag. 40:59–64. 2013.PubMed/NCBI
|
|
19
|
Zhao H, Ma B, Wang Y, Han T, Zheng L, Sun
C, Liu T, Zhang Y, Qiu X and Fan Q: miR-34a inhibits the metastasis
of osteosarcoma cells by repressing the expression of CD44. Oncol
Rep. 29:1027–1036. 2013.PubMed/NCBI
|
|
20
|
Wang Q, Cai J, Wang J, Xiong C and Zhao J:
MiR-143 inhibits EGFR-signaling-dependent osteosarcoma invasion.
Tumour Biol. 35:12743–12748. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Song B, Wang Y, Xi Y, Kudo K, Bruheim S,
Botchkina GI, Gavin E, Wan Y, Formentini A, Kornmann M, et al:
Mechanism of chemoresistance mediated by miR-140 in human
osteosarcoma and colon cancer cells. Oncogene. 28:4065–4074. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhou C, Tan W, Lv H, Gao F and Sun J4:
Hypoxia-inducible microRNA-488 regulates apoptosis by targeting Bim
in osteosarcoma. Cell Oncol (Dordr). 39:463–471. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Lin Z, Song D, Wei H, Yang X, Liu T, Yan W
and Xiao J: TGF-β1-induced miR-202 mediates drug resistance by
inhibiting apoptosis in human osteosarcoma. J Cancer Res Clin
Oncol. 142:239–246. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wang GC, He QY, Tong DK, Wang CF, Liu K,
Ding C, Ji F and Zhang H: MiR-367 negatively regulates apoptosis
induced by adriamycin in osteosarcoma cells by targeting KLF4. J
Bone Oncol. 5:51–56. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhang Y, Duan G and Feng S: MicroRNA-301a
modulates doxorubicin resistance in osteosarcoma cells by targeting
AMP-activated protein kinase alpha 1. Biochem Biophys Res Commun.
459:367–373. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ram Kumar RM, Boro A and Fuchs B:
Involvement and clinical aspects of microRNA in osteosarcoma. Int J
Mol Sci. 17:E8772016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kelly AD, Haibe-Kains B, Janeway KA, Hill
KE, Howe E, Goldsmith J, Kurek K, Perez-Atayde AR, Francoeur N, Fan
JB, et al: MicroRNA paraffin-based studies in osteosarcoma reveal
reproducible independent prognostic profiles at 14q32. Genome Med.
5:22013. View
Article : Google Scholar : PubMed/NCBI
|
|
28
|
Weiner-Gorzel K, Dempsey E, Milewska M,
McGoldrick A, Toh V, Walsh A, Lindsay S, Gubbins L, Cannon A,
Sharpe D, et al: Overexpression of the microRNA miR-433 promotes
resistance to paclitaxel through the induction of cellular
senescence in ovarian cancer cells. Cancer Med. 4:745–758. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Guo LH, Li H, Wang F, Yu J and He JS: The
tumor suppressor roles of mir-433 and mir-127 in gastric cancer.
Int J Mol Sci. 14:14171–14184. 2013. View Article : Google Scholar : PubMed/NCBI
|