Introduction
Glioblastoma (GBM) is an aggressive astrocytic
neoplasm and one of the main contributors to cancer-associated
deaths in adult and pediatric populations (1). Median survival time of GBM patients is
approximately 12–15 months after the first diagnosis (2). Conventional treatments for GBM patients
are mostly surgical tumor excision followed by chemotherapy or
radiation therapy (3). Although the
therapies have increased the survival time to approximately 14.6
months, the advantages are unconspicuous (4). Temozolomide (TMZ) was approved for use
in radiotherapy for treatment of newly diagnosed GBM in adults and
as a maintenance therapy in 2005 (5).
TMZ is the most commonly used and first line chemotherapeutic agent
for GBM patients, though some other chemotherapeutic drugs could be
found as pharmaceutical options (6,7).
Retinoic acid (RA) is a chemotherapeutic and
chemopreventive agent (8). It can
inhibit tumorigenesis via suppressing the cell growth and
stimulating cell differentiation (9).
Besides, RA also promotes cellular apoptosis which may be a reason
of its antitumor characteristics (10). RA mediated signaling influences the
development of the central nervous system through regulating
neuronal differentiation process (11), and at early stages adjusting the
neurogenesis of adults (12).
Autophagy is usually activated in tumors by radiation or
chemotherapy (13,14), and this process acts as a potential
protector for cancer treatment. Autophagy has been thought as a
pro-death or pro-survival response to several external stresses,
such as the radiotherapy and chemotherapy at both organic and
cellular levels (15). The underlying
mechanisms that autophagy has such as seemingly opposite functions
remained elusive. A study reported that autophagy relative pathways
operate to supply cells with metabolic substrates, leading to cell
maintenance in many moderate stimulus conditions (16). However, various studies reported that
autophagy, if uncontrolled, also became a contributor to cell death
that could occur in the absence of detectable apoptosis signs
(17). In addition, one important
autophagy hallmark is the convertion of light chain 3A to its
lapidated form LC3B (18). Besides,
LC3B is the first identified mammalian protein that related to
autophagy (19). Beclin 1 is another
important autophagy- associated protein, which has been reported to
play a central role in neurodegeneration prevention (20).
In the present study, we investigated the the RA
potential therapeutic function to enhance the TMZ effects on the
U251 cells, and explored the growth, proliferation, and apoptosis
of the U251 cells after the RA+TMZ treatment. Moreover, the role of
the RA+TMZ was also elucidated on the regulation of autophagy
associated proteins Beclin 1 and LC3B. Most importantly, the
underlying mechanisms of RA enhancement to TMZ effects on U251
cells were investigated.
Materials and methods
Cell culture
The human U251 glioma cells line was purchased from
the American Type Culture Collection (Rockville, MD, USA). The
cancer cell line was cultured in RPMI-1640 containing 10% fetal
bovine serum (FBS), 50 µg/ml amphotericin B and 100U/ml penicillin,
and maintained at 37°C in 5% CO2 incubator. The control
cells were untreated in medium containing vehicle dimethyl
sulfoxide (DMSO).
MTT assay
Cells were plated in 24-well plates
(5×103 cells per well). TMZ and RA at various
concentrations were added after cells attachment to the plates.
Then after overnight culture, baseline values were obtained by an
MTT assay (Life Technologies, Grand Island, NY, USA). It is a
colorimetric assay to test viable cells by measuring formazan
reduced from MTT. After 20 min of incubation with MTT, we extracted
formazan by DMSO and tested the optical density at 540 nm. MTT
assay was performed at 48 h. Experiments were performed in
triplicate.
Cell cycle analysis by flow
cytometry
U251 cells were collected following the TMZ (400 µM)
and RA (10 µM) and incubated 48 h. Then the cells were fixed in the
ice-cold ethanol (70%) at −20°C overnight. The next morning, the
cells were first incubated in the 10 µM RNAse at 37°C for 30 min,
then stained with propidium iodide at 4°C for 1 h. Cell cycle
analysis was carried out on a FACSCalibur flow cytometer (BD
Biosciences, San Diego, CA, USA) and the obtained data were
analyzed using CellQuest™ software (BD Biosciences).
Cell apoptosis by Annexin V/propidium
iodide (PI) double staining
Following treatment with TMZ (400 µM) and RA (10 µM)
and incubation for 48 h, the cells for apoptosis analysis were
first resuspended in Annexin V binding buffer, and then stained
with the FITC-conjugated Annexin V and PI simultaneously at 37°C
for 15 min in the dark before adding the binding buffer. Then the
apoptotic cells were identified using the flow cytometry analysis
on a FACSCalibur system (FacsScan; BD Biosciences).
Transmission electron microscopy
(TEM)
For the TEM analysis of cell morphological features,
cells were first fixed in 3% glutaraldehyde followed by 1% osmium
tetroxide, then dehydrated in the graded alcohols and embedded in
Epon (Agar Scientific, Stansted, UK). The sections were stained in
lead citrate and uranyl acetate and were finally observed with the
Morgagni 268d transmission electron microscope (FEI, Hillsboro, OR,
USA).
Drug treatment
DMSO, potassium phosphates, Propidium iodide,
Tris-HCl, ribonuclease-A, Triton X-100 and Trypan blue were
obtained from Sigma Chemical Co. (St. Louis, MO, USA). L-glutamine,
DMEM, FBS and penicillin-streptomycin were purchased from Gibco
Invitrogen Corp. (Grand Island, NY, USA). TMZ and RA were obtained
from Sigma Chemical Co. and dissolved in DMSO to the optimal
working concentration.
Western blot analysis
After treatment, the cells were collected and the
protein extracted with RIPA lysis buffer. The protein
concentrations were quantified by the BCA method. Protein samples
(30 µg) were run on 10% gels, and then transferred to the PVDF
membrane. After 1 h of blocking with the 5% non-fat milk, the
membranes were incubated with the primary rabbit anti-LC3B, the
rabbit anti-Beclin 1 antibody, the rabbit anti-Keap1 antibody, the
rabbit anti-Nrf2 antibody, the rabbit anti-ARE antibody (1:1,000;
Abcam, Cambridge, MA, USA) at 4°C overnight. Washing in TBST three
times, the membranes were then incubated with a peroxidase labeled
secondary antibody (1:5,000; Santa Cruz Biotechnology, Inc.,
Dallas, TX, USA) for 2 h. The bands were washed again, enhanced
with chemiluminescence reagents and visualized with the ChemiDoc™
MP Imaging System (Bio-Rad, Berkeley, CA, USA).
Reverse-transcription PCR
After treatment, the cortex was collected to
determine the expression of Keap1/Nrf2/ARE mRNA by RT-PCR. Total
RNA was extracted by the TRIzol RNA extraction reagent (Invitrogen,
Carlsbad, CA, USA) following the protocols and quantified by
spectrophotometer method. Purified RNA with equal volume was
reverse transcribed (RevertAid Fist Strand cDNA Synthesis kit,
K1622; Thermo Fisher Scientific, Waltham, MA, USA). RT-PCR analysis
was performed using a PCR Thermal Cycler Dice instrument (Takara
Bio, Inc., Otsu, Japan).
Statistical analysis
All data were expressed as the mean ± standard error
of the mean (SEM), and were analyzed using a one-way analysis of
variance followed by Bonferroni-Dunn correction. Statistical
analysis was performed using SPSS software, version 20.0 (SPSS,
Inc., Chicago, IL, USA).
Results
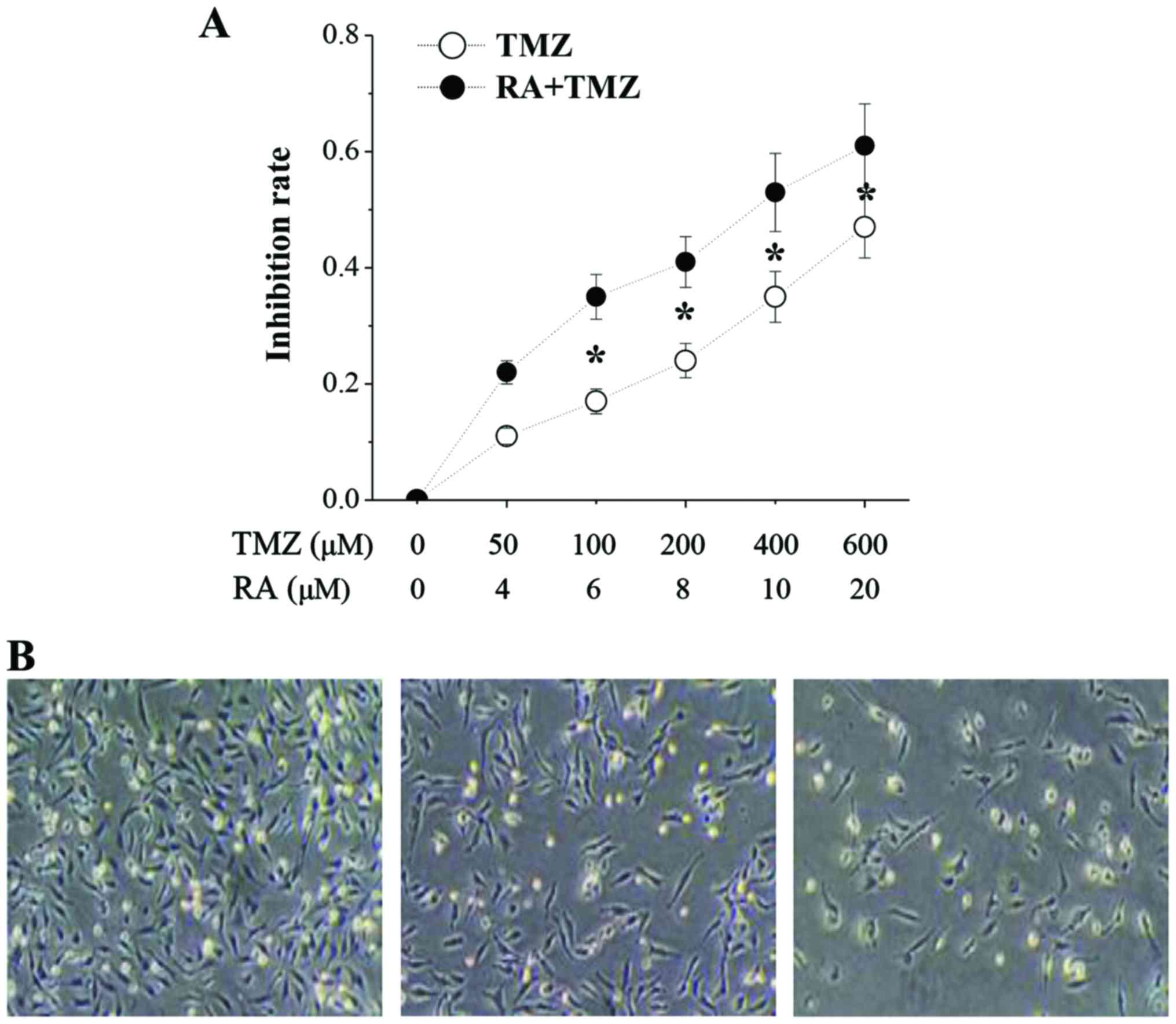
RA enhances the TMZ effects on
inhibition of the U251 cell proliferation
To verify if RA has additive, synergistic or
antagonistic effects after administration with TMZ, U251 cells were
treated with RA and TMZ at various concentrations and incubated for
48 h. As shown in Fig. 1A, the TMZ
IC50 was 691.6 µM while the TMZ+RA IC50 was TMZ 386.7 µM and RA 9.6
µM after incubation for 48 h. As Fig.
2B shown, the MTT assay further conformed that RA enhanced the
TMZ effects on inhibition of the U251 cells proliferation.
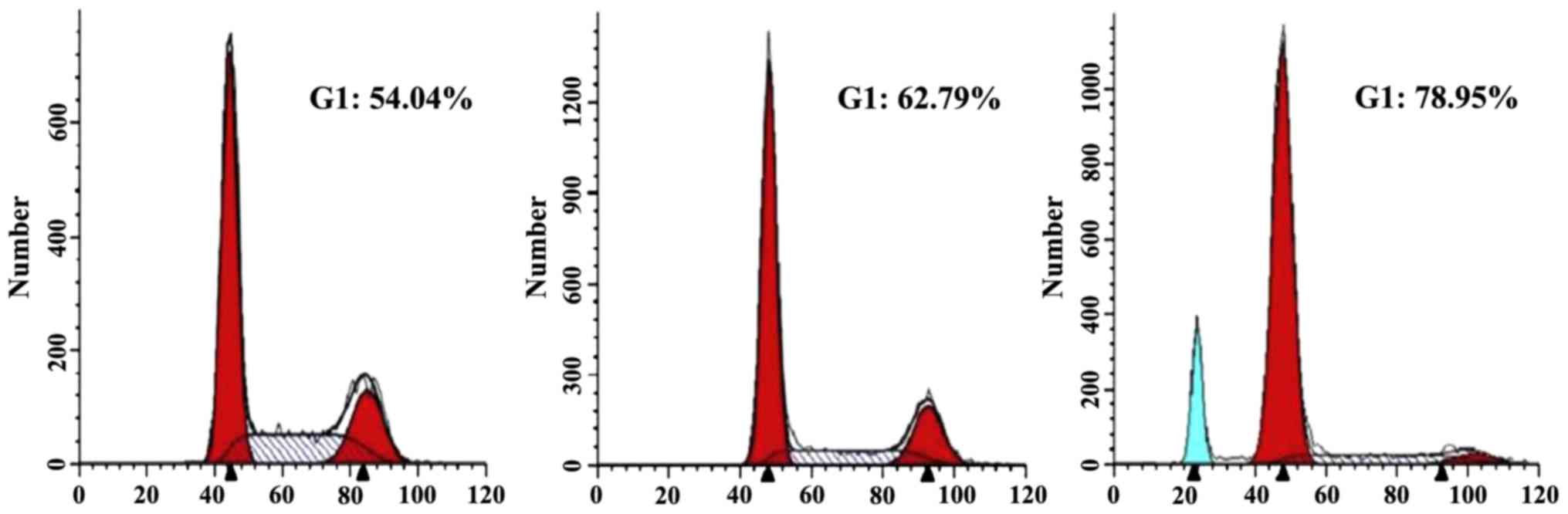
RA enhances the TMZ effects on
modulation of the U251 cell cycle
The U251 cells were treated with RA and TMZ at an
appropriate ratio according to their IC50, respectively, and
incubated for 48 h. Then the flow cytometry analysis was performed
to investigate the U251 cell cycle according to the cell cycle
stage distribution (G0, G1, S, G2 and M). We found a slight G0/G1
phase arrest in the TMZ group and a marked arrest in the RA+TMZ
group, elucidating that RA could enhance the TMZ effects on
modulation of the U251 cell cycle (Fig.
2).
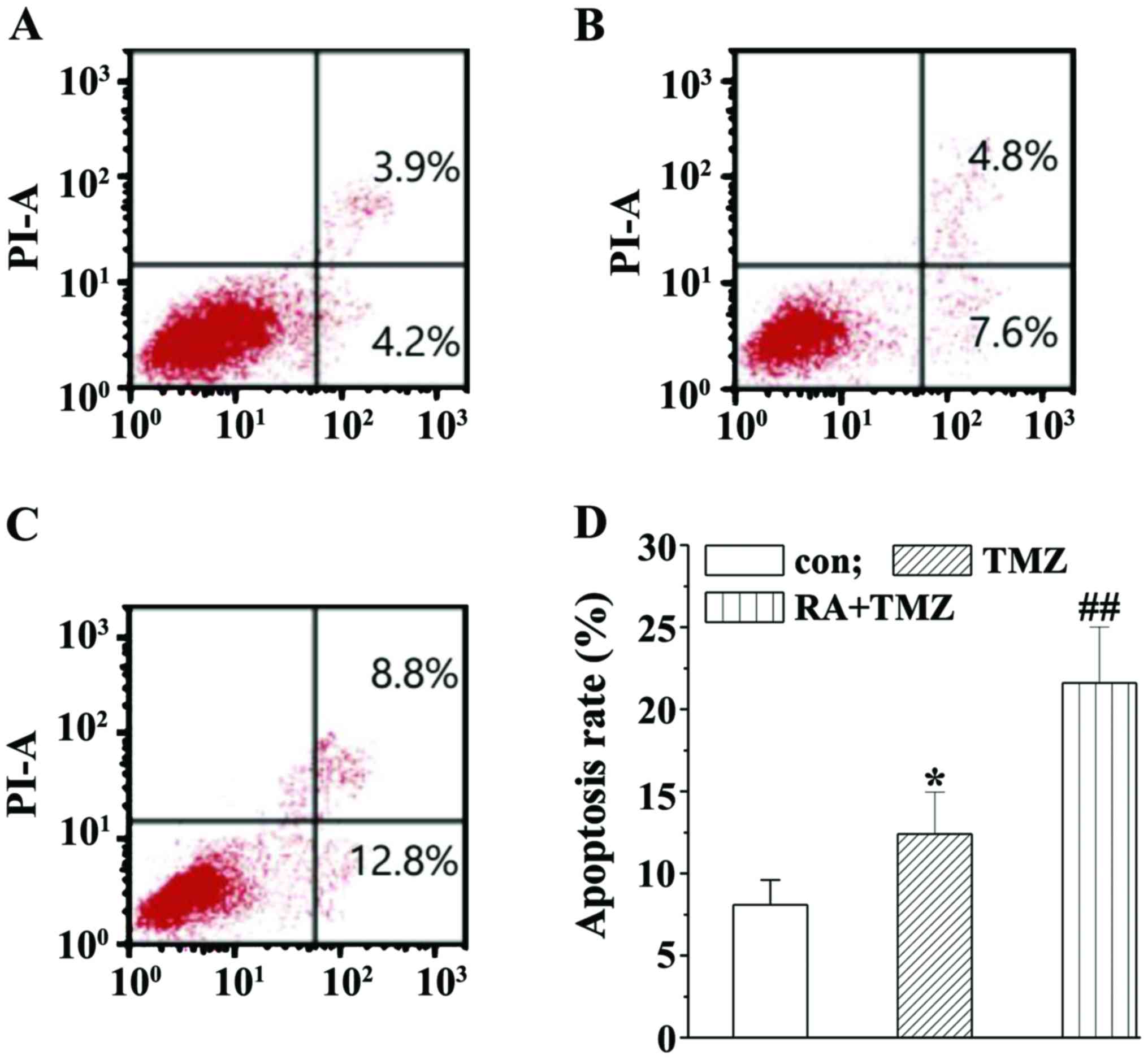
RA enhances the TMZ effects on
induction of U251 cell apoptosis
With the Annexin V/FITC and PI method, the RA
ability to enhance the TMZ effects on inducing U251 cell apoptosis
was explored. U251 cell apoptosis was slightly induced in the TMZ
group while significantly induced by the RA+TMZ treatment (Fig. 3A). As shown in Fig. 3B, the number of apoptotic cells was
markedly increased after treatment with 10 µM RA + 400 µM TMZ
compared with the TMZ group (21.6±1.5% vs. 12.4±1.1%).
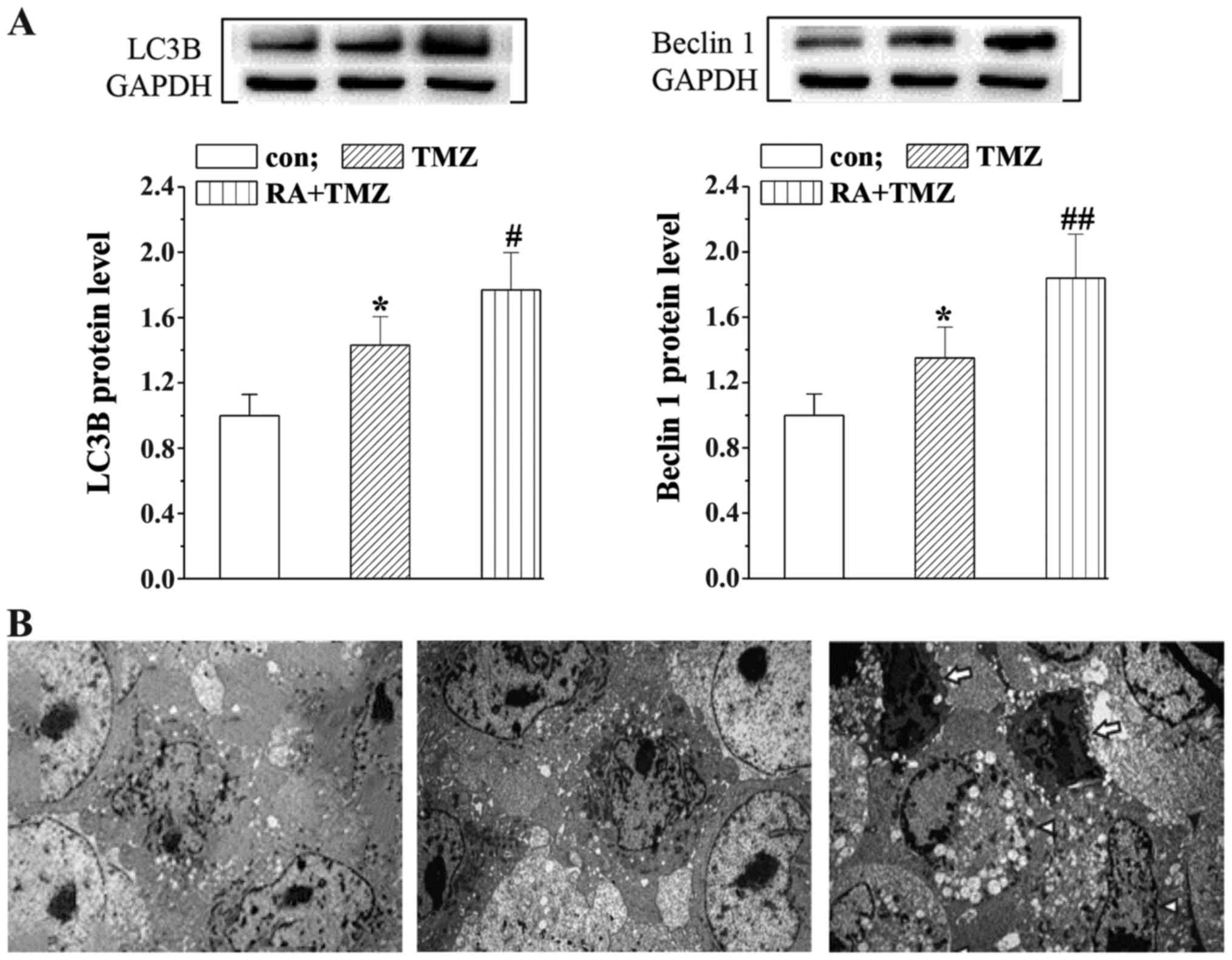
RA enhances the TMZ effects on
induction of U251 cell autophagy
In order to assess the presumed autophagy mechanism
in U251 cells, we detected the protein leves of autophagy
associated proteins Beclin 1 and LC3B with western blot analysis.
The LC3B and Beclin 1 levels were slightly upregualated in the TMZ
group while significantly increased by the RA+TMZ treatment
(Fig. 4A). Furthermore, the results
of ultrastructural TEM analysis were consistent with autophagy
induction, showing an extensive cytoplasm vacuolization and many
single-membrane autophagolysosomes containing cellular debris
(Fig. 4B).
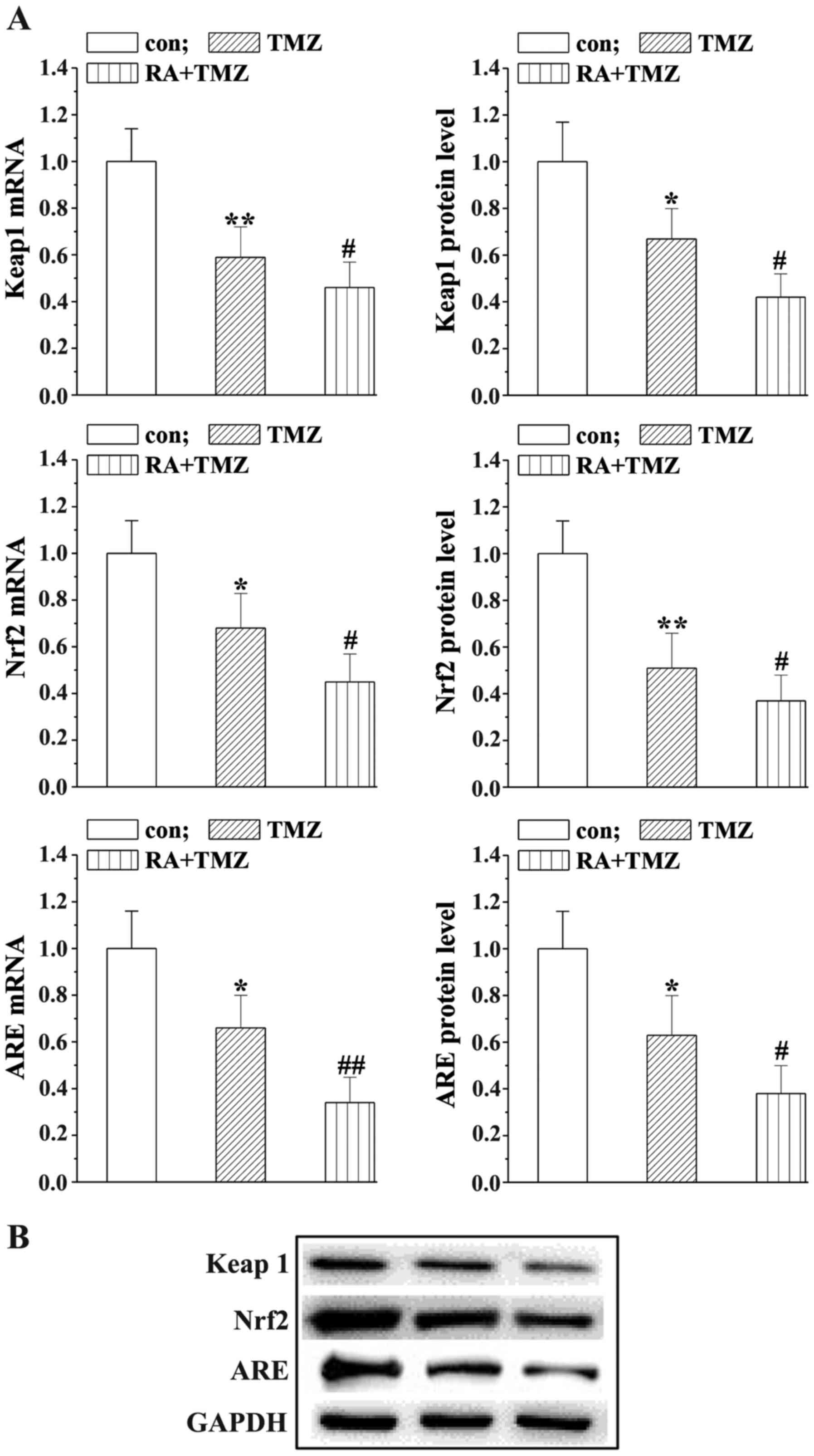
RA enhances the effects of TMZ
reducing the Keap1/Nrf2/ARE signaling
Western blot analysis and RT-PCR were applied to
assess the mRNA and protein levels of Keap1/Nrf2/ARE to explore the
underlying mechanisms of RA in combination with TMZ. As shown in
Fig. 5, the Keap1/Nrf2/ARE expression
was slightly downregulated in the TMZ group while markedly reduced
by the RA+TMZ treatment.
Discussion
In the present study, the RA potential therapeutic
function to enhance the TMZ effects on the U251 cells were
evaluated. Our results suggested that the RA treatment enhanced the
TMZ effects on inhibition of the U251 cell growth and arresting
cell cycle progression at the G0/G1 phase. Besides, RA+TMZ
treatment was able to significantly induce U251 cell apoptosis.
Most importantly, the results of ultrastructural TEM analysis were
consistent with autophagy protein levels, showing an extensive
cytoplasm vacuolization and many single-membrane autophagolysosomes
containing cellular debris. Western blot analysis and RT-PCR
confirmed the downregualtion of Keap1/Nrf2/ARE signaling pathway
involved in the mechanisms that the RA treatment could enhance the
TMZ effects on U251 cells.
In spite of the advances in GBM therapies, the
overall 5 year survival rate is still less than 5% (21). The efficacy of TMZ is often limited
due to the resistance development (22). Studies have found that autophagy
played a central role in the drug treatment of GBM. TMZ induces
autophagy of glioma cells to produce anticancer effect (23,24). It
has been reported that autophagy has close connection with the
chemoresistance in glioma cells (25–27). Even
though autophagy had a suppressive effect on tumor in normal cells,
the autophagy induction may facilitate tumor cells survival,
resulting in the resistance to cellular death. However, autophagy
could also be involved in the death mechanism that inhibited tumor
progression based on the degree of cellular context and the DNA
damage (22). Currently there is no
consistent opinion on whether the autophagic death could be
considered as a form of the programmed cellular death, or whether
it could simply represent dying cells with the autophagic feature
(28). In addition, the neurons and
brain tissues were the major targets for illuminating how RA
deficiency could influence the plasticity, differentiation, and
neuronal neurogenesis (29–31), which may explained the reason for the
altered behavior induced by the RA deficiency. However, the effects
of RA deficiency on the glial cells remain elusive.
The new results in our present study were that the
RA treatment in combination with TMZ in the human U251 cells could
further inhibit the cells growth, arresting cell cycle progression
at the G0/G1 phase, and significantly induced apoptosis of U251
cells. Moreover, after the RA+TMZ treatment of U251 cells,
autophagy-associated proteins Beclin 1 and LC3B were significantly
increased, and the ultrastructural TEM analysis were consistent
with autophagy protein levels. Keap1/Nrf2/ARE expression was
significantly downregulated, indicating the involvement in the
mechanisms that RA treatment could enhance the TMZ effects on U251
cells. The close correlation between the U251 cells and the RA
treatment in combination with TMZ administration may provide some
experimental evidence for the possible effect of the RA+TMZ against
the growth and proliferation of the U251 cells. The clear
mechanisms underlying the RA+TMZ action and its utility for the
treatment of GBM cancer in humans still need to be investigated
further.
References
|
1
|
Hathout L, Ellingson BM, Cloughesy T and
Pope WB: A novel bicompartmental mathematical model of glioblastoma
multiforme. Int J Oncol. 46:825–832. 2015.PubMed/NCBI
|
|
2
|
Huse JT and Holland EC: Targeting brain
cancer: Advances in the molecular pathology of malignant glioma and
medulloblastoma. Nat Rev Cancer. 10:319–331. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bryukhovetskiy IS, Dyuizen IV, Shevchenko
VE, Bryukhovetski AS, Mischenko PV, Milkina EV and Khotimchenko YS:
Hematopoietic stem cells as a tool for the treatment of
glioblastoma multiforme. Mol Med Rep. 14:4511–4520. 2016.PubMed/NCBI
|
|
4
|
Knisely JP and Baehring JM: A silver
lining on the horizon for glioblastoma. Lancet Oncol. 10:434–435.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Nanegrungsunk D, Onchan W, Chattipakorn N
and Chattipakorn SC: Current evidence of temozolomide and
bevacizumab in treatment of gliomas. Neurol Res. 37:167–183. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Messaoudi K, Clavreul A and Lagarce F:
Toward an effective strategy in glioblastoma treatment. Part II:
RNA interference as a promising way to sensitize glioblastomas to
temozolomide. Drug Discov Today. 20:772–779. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wait SD, Prabhu RS, Burri SH, Atkins TG
and Asher AL: Polymeric drug delivery for the treatment of
glioblastoma. Neuro-oncol. 17 Suppl 2:ii9–ii23. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Fu YS, Wang Q, Ma JX, Yang XH, Wu ML,
Zhang KL, Kong QY, Chen XY, Sun Y, Chen NN, et al: CRABP-II
methylation: A critical determinant of retinoic acid resistance of
medulloblastoma cells. Mol Oncol. 6:48–61. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Sakoe Y, Sakoe K, Kirito K, Ozawa K and
Komatsu N: FOXO3A as a key molecule for all-trans retinoic
acid-induced granulocytic differentiation and apoptosis in acute
promyelocytic leukemia. Blood. 115:3787–3795. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Manor D, Shmidt EN, Budhu A,
Flesken-Nikitin A, Zgola M, Page R, Nikitin AY and Noy N: Mammary
carcinoma suppression by cellular retinoic acid binding protein-II.
Cancer Res. 63:4426–4433. 2003.PubMed/NCBI
|
|
11
|
Niederreither K, Abu-Abed S, Schuhbaur B,
Petkovich M, Chambon P and Dollé P: Genetic evidence that oxidative
deriva tives of retinoic acid are not involved in retinoid
signaling during mouse development. Nat Genet. 31:84–88.
2002.PubMed/NCBI
|
|
12
|
Jacobs S, Lie DC, DeCicco KL, Shi Y,
DeLuca LM, Gage FH and Evans RM: Retinoic acid is required early
during adult neurogenesis in the dentate gyrus. Proc Natl Acad Sci
USA. 103:pp. 3902–3907. 2006; View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Sui X, Chen R, Wang Z, Huang Z, Kong N,
Zhang M, Han W, Lou F, Yang J, Zhang Q, et al: Autophagy and
chemotherapy resistance: A promising therapeutic target for cancer
treatment. Cell Death Dis. 4:e8382013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Paglin S, Hollister T, Delohery T, Hackett
N, McMahill M, Sphicas E, Domingo D and Yahalom J: A novel response
of cancer cells to radiation involves autophagy and formation of
acidic vesicles. Cancer Res. 61:439–444. 2001.PubMed/NCBI
|
|
15
|
Fulda S and Kögel D: Cell death by
autophagy: Emerging molecular mechanisms and implications for
cancer therapy. Oncogene. 34:5105–5113. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Galluzzi L, Morselli E, Vicencio JM, Kepp
O, Joza N, Tajeddine N and Kroemer G: Life, death and burial:
Multifaceted impact of autophagy. Biochem Soc Trans. 36:786–790.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Codogno P and Meijer AJ: Autophagy and
signaling: Their role in cell survival and cell death. Cell Death
Differ. 12 Suppl 2:1509–1518. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhang C, Jia X, Wang K, Bao J, Li P, Chen
M, Wan JB, Su H, Mei Z and He C: Polyphyllin VII induces an
autophagic cell death by activation of the JNK pathway and
inhibition of PI3K/AKT/mTOR pathway in HepG2 cells. PLoS One.
11:e01474052016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kabeya Y, Mizushima N, Ueno T, Yamamoto A,
Kirisako T, Noda T, Kominami E, Ohsumi Y and Yoshimori T: LC3, a
mammalian homologue of yeast Apg8p, is localized in autophagosome
membranes after processing. EMBO J. 19:5720–5728. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Cao Y and Klionsky DJ: Physiological
functions of Atg6/Beclin 1: A unique autophagy-related protein.
Cell Res. 17:839–849. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yan Y, Xu Z, Dai S, Qian L, Sun L and Gong
Z: Targeting autophagy to sensitive glioma to temozolomide
treatment. J Exp Clin Cancer Res. 35:232016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhang C, Zhu Q, He H, Jiang L, Qiang Q, Hu
L, Hu G, Jiang Y, Ding X and Lu Y: RIZ1: A potential tumor
suppressor in glioma. BMC Cancer. 15:9902015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhang C, Cai Z, Liang Q, Wang Q, Lu Y, Hu
L and Hu G: RLIP76 depletion enhances autophagic flux in U251
cells. Cell Mol Neurobiol. 37:555–562. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kanzawa T, Germano IM, Komata T, Ito H,
Kondo Y and Kondo S: Role of autophagy in temozolomide-induced
cytotoxicity for malignant glioma cells. Cell Death Differ.
11:448–457. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Shinojima N, Yokoyama T, Kondo Y and Kondo
S: Roles of the Akt/mTOR/p70S6K and ERK1/2 signaling pathways in
curcumin-induced autophagy. Autophagy. 3:635–637. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Natsumeda M, Maitani K, Liu Y, Miyahara H,
Kaur H, Chu Q, Zhang H, Kahlert UD and Eberhart CG: Targeting notch
signaling and autophagy increases cytotoxicity in glioblastoma
neurospheres. Brain Pathol. 26:713–723. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Chen Y, Meng D, Wang H, Sun R, Wang D,
Wang S, Fan J, Zhao Y, Wang J, Yang S, et al: VAMP8 facilitates
cellular proliferation and temozolomide resistance in human glioma
cells. Neuro Oncol. 17:407–418. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Pratt J and Annabi B: Induction of
autophagy biomarker BNIP3 requires a JAK2/STAT3 and MT1-MMP
signaling interplay in Concanavalin-A-activated U87 glioblastoma
cells. Cell Signal. 26:917–924. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Bordin DL, Lima M, Lenz G, Saffi J, Meira
LB, Mésange P, Soares DG, Larsen AK, Escargueil AE and Henriques
JA: DNA alkylation damage and autophagy induction. Mutat Res.
753:91–99. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wagner E, Luo T, Sakai Y, Parada LF and
Dräger UC: Retinoic acid delineates the topography of neuronal
plasticity in postnatal cerebral cortex. Eur J Neurosci.
24:329–340. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Baharvand H, Mehrjardi NZ, Hatami M, Kiani
S, Rao M and Haghighi MM: Neural differentiation from human
embryonic stem cells in a defined adherent culture condition. Int J
Dev Biol. 51:371–378. 2007. View Article : Google Scholar : PubMed/NCBI
|