Introduction
Hepatocellular carcinoma (HCC) represents the third
highest cause of cancer-associated mortality, and one of the most
aggressive human malignancies worldwide (1). Screening of HCC using ultrasonography
and measurement of the serum α-fetoprotein level has allowed early
detection, consequently increasing the survival of patients
(2). However, the overall survival of
patients with HCC following resection remains unsatisfactory due to
recurrence and metastasis. Multiple genetic and epigenetic
alterations occur during the carcinogenesis and progression of HCC
(3–5).
The aggressiveness of this disease may be caused by the activation
of oncogenes, the inactivation of tumor suppressor genes, and the
deregulation of growth factors and their corresponding receptors
(6,7).
However, the mechanism responsible for HCC progression remains
elusive.
Molecular chaperones, a number of which are
heat-shock proteins (HSPs), are often overexpressed in various
types of cancer and have been suggested to be contributing factors
in tumorigenesis (8). Several
previous studies have demonstrated that aberrant expression of
HSP70 members correlates with poor prognosis and resistance to
therapy in malignant human tumors, including breast cancer, bladder
cancer, melanoma and oral cancer (9–12).
Conversely, there are also data that demonstrate that members of
the HSP70 family are involved in antigen processing and
presentation through binding to short peptides, thereby eliciting a
strong antitumor immune response (13). Therefore, the involvement of HSP70 in
such diverse roles may suggest its use in novel anticancer
therapeutic approaches to a broad spectrum of tumor types. Although
HSP70 serves a controversial role in various types of cancer, its
biological effect in HCC remains unknown.
In the present study, the expression of HSP70 and
its function were investigated in HCC cell lines, rendering it a
potential target for therapeutic intervention in HCC.
Materials and methods
Cell culture and reagents
Human normal hepatocyte (L02) and HCC cell lines
(Huh7, HepG2, Hep3B and SMMC-7721) were purchased from the Cell
Bank of Type Culture Collection of Chinese Academy of Sciences
(Shanghai, China). The cells were cultured in a petri-dish
(Corning, Shanghai, China) containing Dulbecco's modified Eagle's
medium (DMEM; Gibco; Thermo Fisher Scientific, Inc.; Waltham, MA,
USA) supplemented with 10% fetal bovine serum (HyClone; GE
Healthcare Life Sciences, Logan, UT, USA) and 1%
penicillin/streptomycin (Gibco; Thermo Fisher Scientific, Inc.), in
an atmosphere containing 5% CO2 at 37°C.
RNA isolation and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted from cells using
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.). The ultramicro spectrophotometer (Thermo Fisher Scientific,
Inc.) was used to detect the purification and quantification of
RNA. Complementary DNA (cDNA) was synthesized using PrimeScript™ RT
reagent kit with gDNA Eraser (Takara Biotechnology Co., Ltd.,
Dalian, China). DNase (Takara Biotechnology Co., Ltd.) was used
prior to the reverse transcription reaction. The conditions for
reverse transcription reaction were as follows: 37°C for 15 min and
then 85°C for 5 sec. RT-qPCR was performed using the Bio-Rad
iCycler iQ Real-Time PCR system (Bio-Rad, Hercules, CA, USA) with
the following primers: HSP70 forward, 5′-ACCTACTCCGACAACCAA-3′ and
reverse, 5′-AGATGACCTCTTGACACTTG-3′; GAPDH forward,
5′-CATGGCAAATTCCATGGCA-3′ and reverse,
5′-TCTAGACGGCAGGTCAGGTCCACC-3′. The PCR mixture was prepared using
SYBR® Master Mix (Takara Biotechnology Co., Ltd.) in
accordance with the protocol of the manufacturer. The conditions
for PCR reaction were as follows, 95°C for 30 sec for one cycle,
and then 95°C for 5 sec, 60°C for 30 sec for 40 cycles. To ensure
that only specific bands were produced, the melting curve was
analyzed at the end of each PCR experiment. Expression levels of
each mRNA were determined using the 2−ΔΔCq method using
GAPDH as an endogenous control (14).
Western blotting
For western blotting, cells were lysed for 30 min on
ice with radioimmunoprecipitation assay buffer containing
phosphatase and protease inhibitors (Beyotime, Haimen, Jiangsu,
China). The cell lysate was then centrifuged at 12,000 × g for 5
min at 4°C. The supernatant was carefully collected following
centrifugation. The protein concentrations were determined using a
bicinchoninic acid protein assay (Pierce; Thermo Fisher Scientific,
Inc.). The cell lysates (50 µg protein/lane) were separated by 10%
SDS-PAGE and transferred to nitrocellulose membranes (HyClone; GE
Healthcare Life Sciences). The membranes were blocked with 5% (v/v)
skim milk at room temperature for 1 h, and then probed with the
primary antibodies at 4°C overnight. Following washing with
TBS/Tween-20 three times, the membranes were incubated with the
horseradish peroxidase-conjugated secondary antibody (cat. no.
7076; Cell Signalling Technology, Danvers, MA, USA; dilution,
1:1,000;) at room temperature for 1 h. A primary antibody against
cyclin-dependent kinase inhibitor 1B (p27Kip1) was purchased from
BD Biosciences (cat. no. 610241; San Jose, CA, USA; dilution,
1:2,000). A primary antibody against Cyclin D1 was purchased from
Santa Cruz Biotechnology Inc. (cat. no. sc-4074; Dallas, TX, USA;
dilution, 1:1,000). Relative optical density of all bands was
analyzed by GelPro Analyzer (V4.0; Media Cybernetics, Rockville,
MD, USA).
Flow cytometry assay
Cell apoptosis was evaluated with the Annexin
V-fluorescein isothiocyanate (FITC)/propidium iodide (PI) apoptosis
detection kit (cat. no. 556547; BD Biosciences) according to the
manufacturer's protocol. Pre-treated Hep3B or SMMC-7721 cells were
stained with FITC-Annexin V and PI, and then evaluated for
apoptosis using flow cytometry (FACSCaliburTM; BD Biosciences),
according to the manufacturer's protocol. Briefly, cells were
harvested following the incubation period, washed twice in cold
PBS, and centrifuged at 300 × g for 5 min at 4°C. A total of ~1×106
cells were re-suspended in 500 µl 1X Annexin-binding buffer and
transferred to a sterile flow cytometry glass tube. A total of 5 µl
FITC-Annexin V and 10 µl PI were added to each tube and incubated
at room temperature for 5 min in the dark. Subsequent to
incubation, samples were analyzed using the standard program of
flow cytometry and analyzed using FlowJo V7.6.1 software (Tree
Star, Inc., Ashland, OR, USA).
Cell proliferation assay
The viability of SMMC-7721 and Hep3B cells
post-transfection was determined using a Cell Counting kit-8
(CCK-8; Dojindo Molecular Technologies, Inc., Kumamoto, Japan).
Cells were seeded in 96-well culture plates at a density of 4×103
cells per well and transfected with the desired small interfering
(si)RNA (GenePharma, Shanghai, China). Cellular transfection was
performed using Lipofectamine 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the protocol. Subsequent to
culturing for 1–5 days at 37°C with 5% CO2, the
supernatant was removed and cell growth was determined using a
CCK-8 kit, according to the manufacturer's protocol. Absorbance was
measured at 450 nm using a microplate reader.
Cell cycle analysis
Cells (1×106 cells/well) were seeded on 6 well
plates (Corning Life Sciences, Shanghai, China) and cultured with
DMEM medium at 37°C with 5% CO2 for 24 h, prior to being
transfected with 100 nM HSP70-siRNA or control siRNA (siCON) for 48
h. For the flow cytometry, cells were trypsinized, pelleted via
centrifugation at 1,000 × g for 5 min at 4°C and resuspended in 300
µl 0.1% Triton X-100 (Sigma-Aldrich; Merck KGaA, Darmstadt,
Germany)/PBS. Cells were fixed with the cold 70% ethanol (Sangon
Biotech Co., Ltd., Shanghai, China) at 4°C for 2 h and centrifuged
at 1,000 × g for 5 min at 4°C. Subsequent to washing twice in cold
PBS and centrifugation at 1,000 × g for 5 min at 4°C, cells were
treated with RNase Type I-A (Sigma-Aldrich; Merck KGaA) at 37°C for
15 min and stained with PI (1 mg/ml) at room temperature for 10 min
in the dark. Cellular DNA content was determined using a
FACSCalibur™ (BD Biosciences). Cell cycle phase distributions were
analyzed by ModFit LT™ (v3.0; BD Biosciences) cell cycle analysis
software.
Cell migration and invasion assay
For the migration assay, 1×104 Hep3B or SMMC-7721
cells were plated into 24-well Boyden chambers (Corning
Incorporated, Corning, NY, USA) with an 8-µm pore polycarbonate
membrane. For the invasion assay, 1×104 Hep3B or SMMC-7721 cells
were plated on chambers pre-coated with 20 µg Matrigel. In these
two assays, the cells were plated in DMEM medium without serum in
the upper chamber, and medium containing 10% fetal bovine serum in
the lower chamber served as a chemoattractant. After 24 h, cotton
swabs removed the cells that did not migrate or invade through the
pores. The inserts were fixed with 4% paraformaldehyde (Sangon,
Shanghai, China) (30 min at room temperature), stained with 0.1%
crystal violet (Beyotime, Haimen, Jiangsu, China) (30 min at room
temperature) and washed with PBS (room temperature). Five random
fields for each insert were counted under an inverted microscope at
×100 magnification (Olympus Corporation, Tokyo, Japan).
Statistical analysis
All statistical analyses were performed using SPSS
v18.0 (SPSS Inc., Chicago, IL, USA). The results are presented as
the means ± standard deviation, and statistical analysis was
performed using the unpaired Student's t-test for two groups and
one-way analysis of variance for more than two groups, followed by
Dunnett's Test for multiple comparisons. P<0.05 was considered
to indicate a statistically significant difference. At least three
replicates were performed for each experiment.
Results
Targeted HSP70 knockdown in HCC cells
by siRNA
To demonstrate the expression of HSP70 in cell
lines, normal hepatocyte L02 and four HCC cell lines (Huh7, HepG2,
SMMC-7721 and Hep3B) were used. In contrast to normal L02
hepatocytes, the four HCC cell lines exhibited greater expression
of HSP70 (Fig. 1A), suggesting that
HSP70 may be involved in hepatocarcinogenesis. Subsequently, to
elucidate the role of HSP70 in hepatocarcinogenesis, the effect of
HSP70 knockdown on HCC cell growth following transfecting HCC cells
with HSP70-siRNA or siCON was examined. In the present study, two
HCC cell lines (SMMC-7721 and Hep3B) were selected as the cell
models, as they expressed higher levels of HSP70 when compared with
the other two cell lines. The results of the western blotting and
qPCR demonstrated that HSP70-siRNA may effectively suppress the
protein and mRNA levels of HSP70 in the two examined cell lines
(Fig. 1B and C).
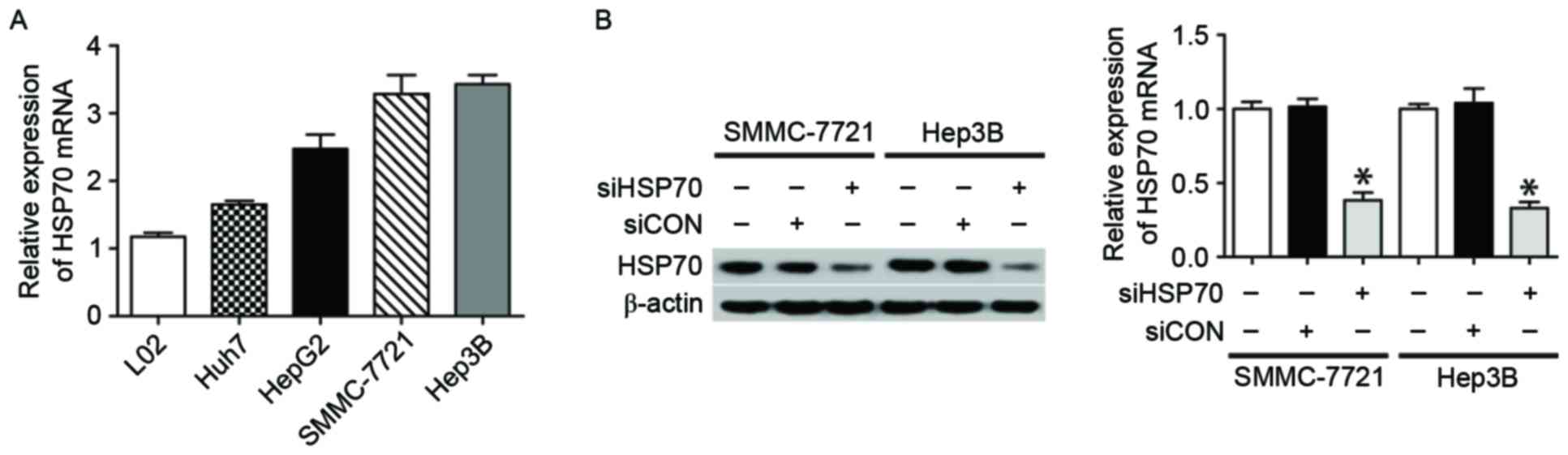 | Figure 1.HSP70 expression in HCC cell lines and
siRNA knockdown of HSP70 expression. (A) qPCR revealed that HSP70
mRNA was expressed in L02 normal hepatocytes and four HCC cell
lines, including Huh7, HepG2, SMMC-7721 and Hep3B. (B) The western
blotting (left) and qPCR (right) results indicate HSP70 expression
in SMMC-7721 and Hep3B cells following various treatments,
including HSP70-siRNA, siCON and no treatment. β-actin served as a
loading control. All data are representative of three independent
experiments. *P<0.05 vs. siHSP70. HSP70, heat shock protein 70;
HCC, hepatocellular carcinoma; si, small interfering; qPCR,
quantitative polymerase chain reaction; CON, control. |
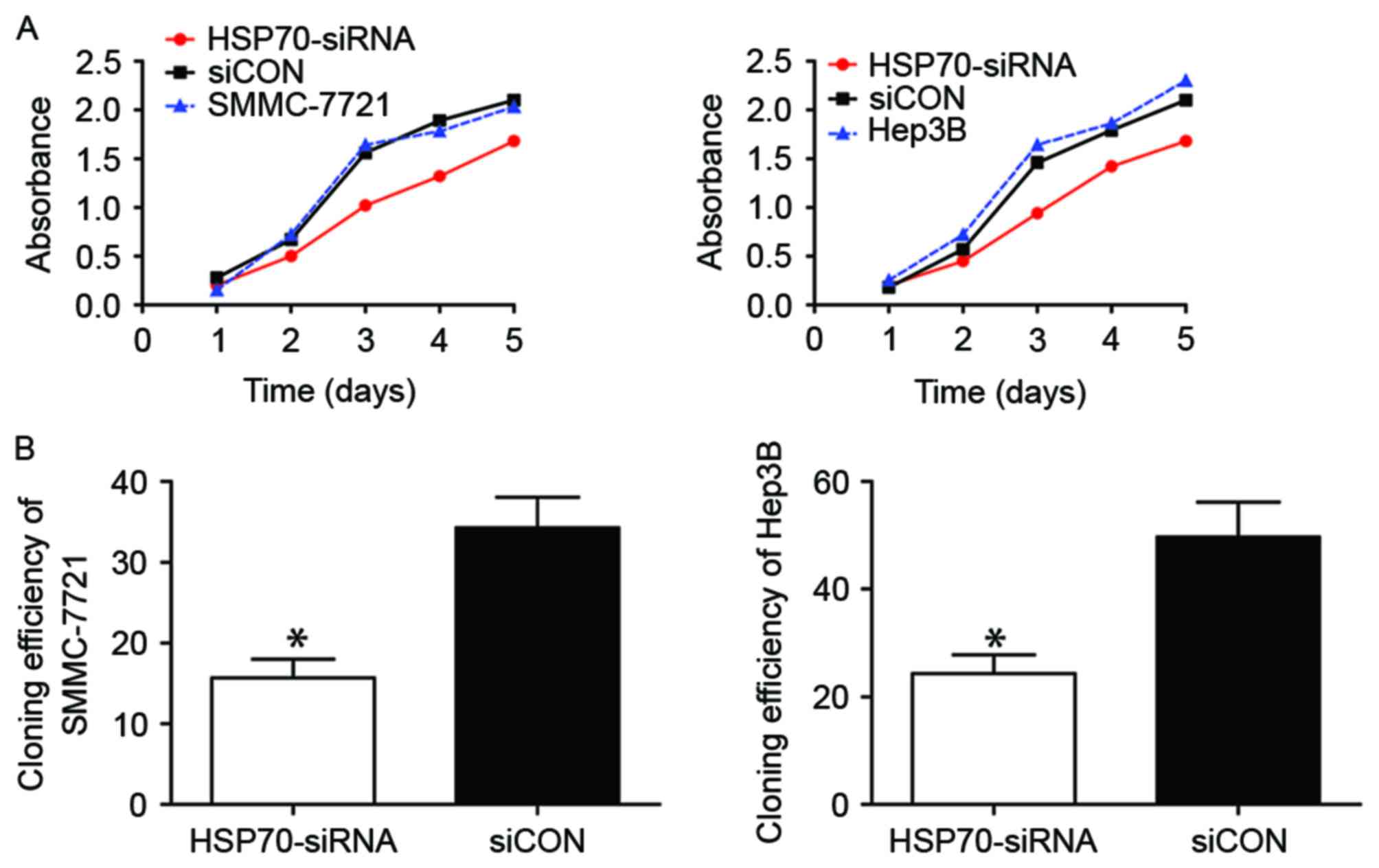
HSP70 knockdown affects the
proliferation ability of HCC cells
The biological role of HSP70 in the growth of HCC
cells was subsequently investigated. The proliferative capacity of
SMMC-7721 (Fig. 2A, left) and Hep3B
(Fig. 2A, right) cells following
HSP70 knockdown decreased markedly when compared with that of
corresponding siCON-transfected control cells, particularly from
day 3–5, as detected using a CCK-8. Furthermore, the cloning
efficiency in the siCON group was significantly greater compared
with that in HSP70-siRNA groups of SMMC-7721 (Fig. 2B, left; P<0.05) and Hep3B (Fig. 2B, left; P<0.05). Therefore,
transfection with HSP70-siRNA markedly reduced the proliferative
ability of the two HCC cell lines, which is concordant with the
aforementioned CCK-8 assay results.
HSP70 expression regulates the
distribution of the cell cycle in HCC cells
To further understand the mechanism underlying
HSP70-knockdown-induced cell growth inhibition, flow cytometry was
performed to examine the cell cycle of SMMC-7721 and Hep3B cells
transfected with HSP70-siRNA or siCON for 48 h. The proportion of
the two cell lines treated with HSP70-siRNA in the G0/G1
stage evidently increased, while those in the S and G2 stages
markedly decreased with respect to the siCON and untreated groups.
In addition, the cell cycle distribution of HCC cells in the siCON
and the untreated group was similar (Fig.
3A and B; P<0.05), and no significant difference was
observed. These results suggested that the growth inhibition
induced by HSP70 downregulation is mediated by cell cycle arrest at
the G1/S phase in HCC cells. Additionally, cyclin D1 and p27Kip1,
two cell cycle-associated molecules, were used to evaluate the
differences in the cell cycle distribution of the two cells. Cyclin
D1 expression was notably decreased, whereas p27Kip1 expression was
markedly increased, in the HSP70-siRNA group, as compared with in
the siCON and untreated groups (Fig.
3C).
HSP70 contributes to migration and
invasion in HCC cells
As recurrence and metastasis are two main factors
contributing to the poor prognosis of HCC, the migratory and
invasive ability of SMMC-7721 and Hep3B cells was examined. The
HSP70-siRNA of SMMC-7721 (Fig. 4A)
and Hep3B (Fig. 4B) cells
significantly inhibited migration compared with control groups
after 48 h. In the Transwell migration and invasion assay (Fig. 4C and D), HSP70 knockdown decreased
cell migration in the two tested cell lines, compared with in the
siCON and control groups. Furthermore, there a higher number of
invaded cells in the siCON and control groups, as compared with
those in the HSP70-siRNA group. Consistently, knockdown of HSP70
inhibited cell migration and invasion. Thus, the data indicate that
an overexpression of HSP70 promotes migration and invasive
capabilities in vitro, and a reduction of HSP70
correspondingly attenuates HCC cell aggression.
Discussion
HSPs were initially identified as proteins that are
significantly induced by heat shock and other chemical and physical
stresses (15). In previous years,
HSPs were demonstrated to be overexpressed in a wide range of human
cancer types, including HCC (16),
and were implicated in tumor cell proliferation, differentiation,
invasion and metastasis (17). HSP70
is a major stress-inducible heat shock protein, which has also been
revealed to be highly elevated in different types of cancer
(18), suggesting that it may also
serve a role in carcinogenesis. As for the role of HSP70 in
regulating the biological activities of HCC, there is only a small
quantity of relevant data available at present. Therefore, a series
of expression and function assays were performed in order to
identify the vital role of HSP70, and to explore the function of
HSP70 in HCC.
In the present study, as demonstrated by qPCR, HSP70
expression was higher in the four types of HCC cell lines compared
with in the normal L02 cells, which is in accordance with in HCC
tissues specimens (19) and other
types of malignant tumors, including lung cancer (20), breast cancer (21) and colorectal carcinoma (22). Subsequently, SMMC-7721 and Hep3B were
selected for additional study as HSP70 expression was higher in
these two HCC cell lines, compared with the other two HCC cell
lines. As western blotting and qPCR revealed, HSP70-siRNA markedly
reduced HSP70 expression in the SMMC-7721 and Hep3B cells.
Next, the proliferation assay indicated that
transfection with HSP70-siRNA markedly decreased the proliferation
potential of SMMC-7721 and Hep3B cells when compared with the siCON
and untreated groups. A potential explanation is that the knockdown
of HSP70 may inhibit Wnt/β-catenin signaling, which serves a
pivotal role in the progression of HCC (23,24). This
difference was significant, particularly from days 3–5 following
transfection; this is concordant with the western blot analysis
results, as the knockdown of HSP70 expression was most notable
following transfection for 48 h. Furthermore, the cloning
efficiency in the HSP70-siRNA group was markedly lower compared
with in the siCON group, which was also in accordance with the
CCK-8 assay results. Therefore, knockdown of HSP70 inhibited the
proliferation ability of the two HCC cell lines.
Following this, flow cytometry assays demonstrated
that the growth inhibitory effect of HSP70 was caused by cell cycle
arrest at G1/S phase. The expression of the cell cycle
regulator cyclin D1 was markedly downregulated, while the
expression of p27Kip1 was upregulated. Cyclin D1 is
considered to be involved in altering cell cycle progression and is
a downstream target of Wnt/β-catenin signaling (25). It is frequently observed in a variety
of tumor types, and may contribute to tumorigenesis (26,27). In
addition, p27Kip1, a cell-cycle inhibitory molecule,
suppresses the catalytic activity of cyclin D-cyclin-dependent
kinase 4 (28), and is overexpressed
in the very early stages of HCC development (29). Ray et al (28) demonstrated that HSP70 is strictly
regulated during the cell cycle and that levels of HSP70 mRNA
rapidly increase 10–15-fold upon entry into the S phase, and then
decline by the late S and G2 phase. Therefore, the
knockdown of HSP70 may inhibit the Wnt/β-catenin signaling pathway,
contributing to the downregulation of target molecule Cyclin D1 and
an inhibition of the cell cycle. In the present study, the
population of the two HCC cell lines (SMMC-7721 and Hep3B) treated
with HSP70-siRNA in G0/G1 stage markedly
increased, while those in S and G2/M stage markedly
decreased compared with the siCON and untreated groups, which
indicated that the cell cycle was activated by HSP70 and inhibited
by HSP70-siRNA at the G1/S stage.
Invasion and metastasis are two of the principal
hallmarks of cancer and are associated with malignant
cancer-associated mortality, particularly for HCC. The long-term
survival of patients with HCC following curative resection retains
the obstacle of a high recurrence rate, which is primarily due to
the spread of intrahepatic metastases (30). Therefore, the identification of
metastatic factors and an understanding of the underlying molecular
pathways that are involved in the progression of metastasis are
critical. Previous studies have demonstrated that HSPs serve a key
role in the invasion and metastasis of various tumors, thereby
opening a novel avenue for investigating the molecular mechanisms
of tumor progression and develop potential therapeutics (31). A number of studies have implicated
HSP70 as an important regulator for multiple steps of metastasis in
human cancer (17,32,33). Fang
et al (34) indicated that
HSF1 was capable of promoting HCC cell migration and invasion by
facilitating the expression and phosphorylation of HSP27. Hartmann
et al (35) suggested that
Hsp90 inhibition decreases the migration and invasion abilities of
lung carcinoma and glioblastoma cell lines. In the present study,
HSP70 knockdown was demonstrated to markedly inhibit HCC cell
invasion and metastasis in the two cell lines; however, the
potential underlying mechanism requires additional exploration.
In conclusion, the present study provides an early
analysis of the pivotal role of HSP70 in the tumorigenesis and
progression of HCC. These data demonstrate that the knockdown of
HSP70 expression leads to decreasing proliferative ability,
inhibition of the cell cycle and the suppression of invasive and
metastatic capacity in two HCC cell lines with relatively lower
HSP70 expression levels. Taking these results into consideration,
along with those from previous studies, the observations from the
present study suggest that HSP70 may be a promising target for
future therapy for HCC.
Acknowledgements
The present study was supported by a grant from the
International Cooperative Projects of Hainan Province (grant no.
KJHZ2014-14).
Glossary
Abbreviations
Abbreviations:
|
HCC
|
hepatocellular carcinoma
|
|
HSP
|
heat-shock protein
|
References
|
1
|
Llovet JM, Ricci S, Mazzaferro V, Hilgard
P, Gane E, Blanc JF, de Oliveira AC, Santoro A, Raoul JL, Forner A,
et al: Sorafenib in advanced hepatocellular carcinoma. N Engl J
Med. 359:378–390. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Yin J, Zhu JM and Shen XZ: The role and
therapeutic implications of RING-finger E3 ubiquitin ligases in
hepatocellular carcinoma. Int J Cancer. 136:249–257. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Hanahan D and Weinberg RA: Hallmarks of
cancer: The next generation. Cell. 144:646–674. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Yasui W, Sentani K, Sakamoto N, Anami K,
Naito Y and Oue N: Molecular pathology of gastric cancer: Research
and practice. Pathol Res Pract. 207:608–612. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Dawson MA and Kouzarides T: Cancer
epigenetics: From mechanism to therapy. Cell. 150:12–27. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Farazi PA and DePinho RA: Hepatocellular
carcinoma pathogenesis: From genes to environment. Nat Rev Cancer.
6:674–687. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
7
|
Shi X, Zhu HR, Liu TT, Shen XZ and Zhu JM:
The Hippo pathway in hepatocellular carcinoma: Non-coding RNAs in
action. Cancer Lett. 400:175–182. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Mosser DD and Morimoto RI: Molecular
chaperones and the stress of oncogenesis. Oncogene. 23:2907–2918.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Vargas-Roig LM, Fanelli MA, López LA, Gago
FE, Tello O, Aznar JC and Ciocca DR: Heat shock proteins and cell
proliferation in human breast cancer biopsy samples. Cancer Detect
Prev. 21:441–451. 1997.PubMed/NCBI
|
|
10
|
Lazaris AC, Theodoropoulos GE, Aroni K,
Saetta A and Davaris PS: Immunohistochemical expression of C-myc
oncogene, heat shock protein 70 and HLA-DR molecules in malignant
cutaneous melanoma. Virchows Arch. 426:461–467. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kaur J, Srivastava A and Ralhan R:
Expression of 70-kDa heat shock protein in oral lesions: Marker of
biological stress or pathogenicity. Oral Oncol. 34:496–501. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Syrigos KN, Harrington KJ, Karayiannakis
AJ, Sekara E, Chatziyianni E, Syrigou EI and Waxman J: Clinical
significance of heat shock protein-70 expression in bladder cancer.
Urology. 61:677–680. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Udono H and Srivastava PK: Heat shock
protein 70-associated peptides elicit specific cancer immunity. J
Exp Med. 178:1391–1396. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Lindquist S and Craig EA: The heat-shock
proteins. Annu Rev Genet. 22:631–677. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lim SO, Park SG, Yoo JH, Park YM, Kim HJ,
Jang KT, Cho JW, Yoo BC, Jung GH and Park CK: Expression of heat
shock proteins (HSP27, HSP60, HSP70, HSP90, GRP78, GRP94) in
hepatitis B virus-related hepatocellular carcinomas and dysplastic
nodules. World J Gastroenterol. 11:2072–2079. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ciocca DR, Green S, Elledge RM, Clark GM,
Pugh R, Ravdin P, Lew D, Martino S and Osborne CK: Heat shock
proteins hsp27 and hsp70: Lack of correlation with response to
tamoxifen and clinical course of disease in estrogen
receptor-positive metastatic breast cancer (a Southwest Oncology
Group Study). Clin Cancer Res. 4:1263–1266. 1998.PubMed/NCBI
|
|
18
|
Nylandsted J, Brand K and Jäättelä M: Heat
shock protein 70 is required for the survival of cancer cells. Ann
N Y Acad Sci. 926:122–125. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Murphy ME: The HSP70 family and cancer.
Carcinogenesis. 34:1181–1188. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Malusecka E, Zborek A, Krzyzowska-Gruca S
and Krawczyk Z: Expression of heat shock proteins HSP70 and HSP27
in primary non-small cell lung carcinomas. An immunohistochemical
study. Anticancer Res. 21:1015–1021. 2001.PubMed/NCBI
|
|
21
|
Lazaris ACh, Chatzigianni EB,
Panoussopoulos D, Tzimas GN, Davaris PS and Golematis BCh:
Proliferating cell nuclear antigen and heat shock protein 70
immunolocalization in invasive ductal breast cancer not otherwise
specified. Breast Cancer Res Treat. 43:43–51. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Hwang TS, Han HS, Choi HK, Lee YJ, Kim YJ,
Han MY and Park YM: Differential, stage-dependent expression of
Hsp70, Hsp110 and Bcl-2 in colorectal cancer. J Gastroenterol
Hepatol. 18:690–700. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Thompson MD and Monga SP: WNT/beta-catenin
signaling in liver health and disease. Hepatology. 45:1298–1305.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wands JR and Kim M: WNT/beta-catenin
signaling and hepatocellular carcinoma. Hepatology. 60:452–454.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Botrugno OA, Fayard E, Annicotte JS, Haby
C, Brennan T, Wendling O, Tanaka T, Kodama T, Thomas W, Auwerx J
and Schoonjans K: Synergy between LRH-1 and beta-catenin induces G1
cyclin-mediated cell proliferation. Mol Cell. 15:499–509. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Tetsu O and McCormick F: Beta-catenin
regulates expression of cyclin D1 in colon carcinoma cells. Nature.
398:422–426. 1999. View
Article : Google Scholar : PubMed/NCBI
|
|
27
|
Buckley MF, Sweeney KJ, Hamilton JA, Sini
RL, Manning DL, Nicholson RI, deFazio A, Watts CK, Musgrove EA and
Sutherland RL: Expression and amplification of cyclin genes in
human breast cancer. Oncogene. 8:2127–2133. 1993.PubMed/NCBI
|
|
28
|
Ray A, James MK, Larochelle S, Fisher RP
and Blain SW: p27Kip1 inhibits cyclin D-cyclin-dependent kinase 4
by two independent modes. Mol Cell Biol. 29:986–999. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Yachida S, Sakamoto M, Imaida K, Yokohira
M, Saoo K, Okano K, Wakabayashi H, Maeta H and Suzuki Y:
p27(Kip1)is overexpressed in very early stages of
hepatocarcinogenesis. Cancer Sci. 99:2152–2159. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Poon RT, Fan ST, Ng IO, Lo CM, Liu CL and
Wong J: Different risk factors and prognosis for early and late
intrahepatic recurrence after resection of hepatocellular
carcinoma. Cancer. 89:500–507. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Calderwood SK, Khaleque MA, Sawyer DB and
Ciocca DR: Heat shock proteins in cancer: Chaperones of
tumorigenesis. Trends Biochem Sci. 31:164–172. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Karlsson R, Pedersen ED, Wang Z and
Brakebusch C: Rho GTPase function in tumorigenesis. Biochim Biophys
Acta. 1796:91–98. 2009.PubMed/NCBI
|
|
33
|
Nikfarjam M, Muralidharan V, Su K,
Malcontenti-Wilson C and Christophi C: Patterns of heat shock
protein (HSP70) expression and Kupffer cell activity following
thermal ablation of liver and colorectal liver metastases. Int J
Hyperthermia. 21:319–332. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Fang F, Chang R and Yang L: Heat shock
factor 1 promotes invasion and metastasis of hepatocellular
carcinoma in vitro and in vivo. Cancer. 118:1782–1794. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Hartmann S, Günther N, Biehl M, Katzer A,
Kuger S, Worschech E, Sukhorukov VL, Krohne G, Zimmermann H,
Flentje M and Djuzenova CS: Hsp90 inhibition by NVP-AUY922 and
NVP-BEP800 decreases migration and invasion of irradiated normoxic
and hypoxic tumor cell lines. Cancer Lett. 331:200–210. 2013.
View Article : Google Scholar : PubMed/NCBI
|