Introduction
Lung cancer is the leading cause of
cancer-associated mortality worldwide. Non-small cell lung cancer
(NSCLC) accounts for ~85% of all lung cancer cases (1). The prognosis for patients with NSCLC
remains poor, with a 5-year survival rate of ~17% (2). The low survival rate of NSCLC is
primarily due to tumor cell metastasis (3). Therefore, it is necessary to investigate
the mechanisms underlying NSCLC metastasis.
MicroRNAs (miRNAs) are 19–24 nucleotides small
non-coding RNAs, which directly bind to target motifs in mRNAs and
post-transcriptionally suppress gene expression by transcript
degradation or translational repression (4,5). There is
growing evidence that miRNA dysfunction is involved in the growth
and/or metastasis of various types of human tumors (6,7). MicroRNA
(miR)-124, a widely studied miRNA, was demonstrated to be
downregulated and regarded as a tumor suppressor in breast, gastric
and bladder cancer, as well as head and neck squamous cell
carcinoma (8–11). Xi et al (12) and Sun et al (13) recently demonstrated that miR-124
significantly repressed cell invasion and metastasis in colorectal
cancer and NSCLC. Decreased expression of miR-124 was associated
with poor prognosis in patients with breast cancer or NSCLC
(14,15). These results suggested that miR-124
may serve an important role in the regulation of tumor metastasis.
Although miR-124 may inhibit NSCLC metastasis by targeting MYO10
(13), the other targets of miR-124
in this process cannot be excluded.
LIM-homeobox domain 2 (LHX2), a member of the
LIM-homeodomain proteins, was previously reported to serve an
important role in the control of lymphoid and neural cell
differentiation and brain and eye development (16). LHX2 was also implicated in the
development of various types of human tumors. For example, LHX2 may
promote breast cancer cell growth and metastasis by stimulating the
activity of platelet-derived growth factor subunit B signaling
pathway (17). The authors of the
present study previously demonstrated that LHX2 was highly
expressed and may serve an oncogenic role in NSCLC (18). Although data of the previous study
demonstrated that knockdown of LHX2 inhibited NSCLC cell
proliferation and arrested cell cycle at G1 phase
(18), it remains unclear whether
LHX2 affects the migratory and invasive abilities of NSCLC
cells.
Low miR-124and high LHX2 expression levels have been
observed in different cancer types in humans. Therefore, there may
be a link between miR-124 and LHX2 in NSCLC. In order to
investigate this hypothesis, the present study first used
TargetScanHuman v7.0 software to predict miRNA targets and
demonstrated that the 3′-untranslated region (3′-UTR) of the LHX2
transcript was a putative target of miR-124. therefore, this
attracted our attention to the association between miR-124 and LHX2
in NSCLC.
To the best of our knowledge, the present study is
the first time that the role of LHX2 in NSCLC cell invasion and an
association between miR-124 and LHX2 in NSCLC has been
investigated. The results revealed that LHX2 has an important role
in promoting NSCLC cell migration and invasion, which maybe
controlled at least partially by miR-124.
Materials and methods
Cell culture
Human bronchial epithelial (HBE) cells (Bogoo
Biotechnology, Shanghai, China) and human NSCLC cells A549,
LTEP-a2, H1299 (two lung adenocarcinoma cell lines), H226 (lung
squamous carcinoma cell line), 95C and 95D (two giant-cell
carcinoma cell lines) and H460 (large cell carcinoma cell line)
from the Cell Bank of the Chinese Academy of Sciences (Shanghai,
China), were cultured in RPMI-1640 medium (HyClone, Logan, UT, USA)
supplemented with 10% fetal bovine serum (FBS; Invitrogen; Thermo
Fisher Scientific, Inc., Waltham, MA, USA), L-glutamine and 50 U/ml
each of penicillin and streptomycin (Invitrogen; Thermo Fisher
Scientific, Inc.) at 37°C with 5% CO2 in a humidified
atmosphere.
Tissue samples
A total of 40 paired tumor tissues and adjacent
noncancerous tissues were collected, by surgical resection, from
patients with NSCLC at the First Affiliated Hospital of Soochow
University (Suzhou, China) between April 2007 and December 2013.
The demographic and clinical features were described in Table II. Written informed consent was
obtained from all patients prior to enrollment in the present
study. Histological and pathological diagnostics for patients with
NSCLC were evaluated according to the Revised International System
for Staging Lung Cancer (19). None
of the patients received chemotherapy or radiotherapy prior to
tissue sampling. The samples were snap-frozen in liquid nitrogen
and stored at −80°C. The present study was approved by the Academic
Advisory Board of Soochow University.
 | Table II.Comparison of various
clinicopathological parameters with LHX2 mRNA and miR-124
expression in 40 NSCLC samples. |
Table II.
Comparison of various
clinicopathological parameters with LHX2 mRNA and miR-124
expression in 40 NSCLC samples.
|
|
| Ratio of expression
(T/N) |
|---|
|
|
|
|
|---|
| Parameter | n | LHX2 mRNA | miR-124 |
|---|
| Age |
|
|
|
| ≤65 | 18 | 20.61±7.69 | 0.73±0.19 |
|
>65 | 22 | 7.41±2.55 | 0.68±0.22 |
| aP-value |
| 0.086 | 0.864 |
| Sex |
|
|
|
|
Male | 28 | 13.56±4.96 | 0.67±0.20 |
|
Female | 12 | 12.80±5.67 | 6.03±5.23 |
| aP-value |
| 0.929 | 0.144 |
| Smoking status |
|
|
|
| No | 20 | 12.13±3.91 | 3.82±3.15 |
|
Yes | 20 | 14.54±6.68 | 0.73±0.27 |
| aP-value |
| 0.892 | 0.298 |
| Lymph node
metastasis |
| No | 20 | 14.13±6.67 | 3.76±3.15 |
|
Yes | 20 | 12.54±3.95 | 0.79±0.27 |
| aP-value |
| 0.267 | 0.543 |
| Histology |
|
|
|
|
Adenocarcinoma | 23 | 10.16±3.71 | 3.38±2.74 |
|
Squamous cell carcinoma | 14 | 18.37±8.82 | 0.89±0.37 |
|
Others | 3 | 14.17±13.59 | 0.29±0.12 |
| bP-value |
| 0.205 | 0.669 |
RNA extraction, cDNA synthesis and
reverse transcription-quantitative polymerase chain reaction
(RT-qPCR)
Total RNA was isolated from NSCLC cells and human
NSCLC tissues using the HP Total RNA kit (Omega Bio-Tek, Inc.,
Norcross, GA, USA). Purified RNA was reversed transcribed to cDNA
using the M-MLV cDNA Reverse Transcription kit (Invitrogen; Thermo
Fisher Scientific, Inc.), and the sequences of the primers used are
presented in Table I. RT-PCR was
performed using a Platinum SYBR Green qPCR SuperMix-UDG kit (Thermo
Fisher Scientific, Inc.), according to the manufacturer's protocol
on Roche Lightcycler 96 (Roche Diagnostics, Basel, Switzerland).
The thermocycling conditions for qPCR were as follows: 95°C for 5
min, 40 cycles of 95°C for 10 sec, 60°C for 30 sec, followed by
72°C for 10 min. Each RT-qPCR experiment was performed in
triplicate. The relative expression levels of LHX2 mRNA and miR-124
were normalized to β-actin mRNA and U6, respectively, and evaluated
according to the 2−ΔΔCt method (20).
 | Table I.Primers for reverse transcription or
amplification of miR-124 and LHX2 mRNA. |
Table I.
Primers for reverse transcription or
amplification of miR-124 and LHX2 mRNA.
| Primer | Sequence (5′-3′) |
|---|
| RT |
|
| U6 |
CGAGCACAGAATCGCTTCACGAATTTGCGTGTCAT |
|
miR-124 |
GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACGGCATT |
| qPCR |
|
| U6,
forward |
CGAGCACAGAATCGCTTCA |
| U6,
reverse |
CTCGCTTCGGCAGCACATAT |
| miR-124,
forward |
GTGCAGGGTCCGAGGTATT |
| miR-124,
reverse |
GCTAATAAGGCACGCGGTG |
| β-actin,
forward |
CACAGAGCCTCGCCTTTGCC; |
| β-actin,
reverse |
ACCCATGCCCACCATCACG |
| LHX2,
forward |
TTCCAGAACGCCCGAGCCAA |
| LHX2,
reverse |
GGGGCTAGTCAAGTCTGTC |
Western blot analysis
Cells and tissues were lysed and subjected to
western blot analysis as previously described (7). Antibodies applied in the analysis were
as follows: rabbit anti-LHX2 (catalog no. sc-367972; dilution,
1:1,000; Santa Cruz Biotechnology, Inc., Dallas, TX, USA) and mouse
anti-β-actin (catalog no. CW0096M; dilution, 1:2,000; CWBIO,
Beijing, China) primary antibodies, and horseradish peroxidase
(HRP)-conjugated goat anti-rabbit (catalog no. CW0103S; dilution,
1:3,000; CWBIO) or HRP-conjugated goat anti-mouse secondary
antibodies (catalog no. CW0107S; dilution, 1:3,000; CWBIO). The
LHX2 expression level was normalized to β-actin.
Plasmid construction and luciferase
assay
A 72 base pair (bp) DNA sequence of LHX2 3′-UTR
containing a predicted miR-124 target site (position 315–312,
predicted using TargetScanHuman software; version 7.0; www.targetscan.org) was directly synthesized (GENEWIZ,
Suzhou, China) and subcloned into a dual-luciferase report vector,
psiCHECK-2 (Promega Corporation, Madison, WI, USA), in order to
generate a psiCHECK-2-LHX2 3′-UTR-wildtype. In addition, another
similar 72 bp DNA fragment containing a mutant miR-124 target site
was synthesized for construction of a psiCHECK-2-LHX2 3′UTR-mutant
vector (GENEWIZ). Subsequently, each of the luciferase constructs
was co-transfected with miR-124 mimics (5-UAA GGC ACG CGG UGA AUG
CC-3′) or a scrambled negative control (miR-NC;
5′-UUCUCCGAACGUGUCACGUTT-3′) into A549 and H1299 cells,
respectively. Following transfection for 48 h at 37°C, the cells
were lysed and analyzed for luciferase activities using the
dual-luciferase reporter assay system (Promega Corporation). Each
experiment was performed in triplicate. Results are represented as
relative to Renilla luciferase activities, which were normalized to
firefly luciferase activities. All transient transfections,
including anti-miR-124 (5′-GGCAUUCACCGCGUGCCUUA-3′) and anti-miR-NC
(5′-CAGUACUUUUGUGUAGUACAA-3′) were performed using Lipofectamine
2000 (Thermo Fisher Scientific, Inc.), according to the
manufacturer's instructions.
Transient RNA interference
Two short interfering RNAs (siRNA), which
specifically target the LHX2 transcript were directly synthesized
(GenePharma Co., Ltd., Shanghai, China). Sequences for LHX2 siRNAs
were as follows: si-LHX2-1, 5′-GCTTCGGACCATGAAGTCTTA-3′; si-LHX2-2,
5′-GCAACCTCTTACGGCAGGAAA-3′. A scrambled sequence
(5′-TTCTCCGAACGTGTCACGT-3′) was used as the negative control
(si-NC). The cells were transiently transfected with 100 pmol of
siRNA or si-NC for 48 h at 37°C, using Lipofectamine 2000 (Thermo
Fisher Scientific, Inc.). Following 3 days, the cells were
collected for further experiments.
Transwell migration and Matrigel
invasion assays
Transwell migration and Matrigel invasion assays
were performed using Transwell plates with 0.8 µm pore
polycarbonate membranes (BD Biosciences, Franklin Lakes, NJ, USA).
A549 and H1299 cells (5×104) supplemented with 1% FBS
were seeded in the upper chamber [for the invasion assay, Matrigel
(Corning, NY, USA) was added in the upper chamber prior to cell
seeding] and allowed to invade to the reverse side of the chamber
under chemoattractant conditions with 10% FBS medium in the lower
chamber. Following incubation for 48 h at 37°C, the cells on the
upper side were wiped, and invaded cells on the lower side were
stained with 1% crystal violet (BioTime Inc., Alameda, CA, USA) and
imaged and counted under three microscopic fields (light
microscope; magnification, ×100).
Statistical analysis
All statistical analyses were performed using
GraphPad Prism version 5.02 software (GraphPad Software, Inc., La
Jolla, CA, USA). Differences in LHX2 and miR-124 expression levels
between NSCLC tissues (T) and adjacent noncancerous lung
tissues(N)were analyzed using a paired t-test (two-tailed), and
data are presented as the mean ± standard error. Comparisons
between clinicopathological characteristics and expression ratios
(T/N) of LHX2 and miR-124 in NSCLC tissues were performed using
nonparametric tests (Mann-Whitney U test for 2 groups,
Kruskal-Wallis test for ≥3 groups). For cell lines, differences
between 2 groups were assessed using an unpaired t-test
(two-tailed), and data are presented as the mean ± standard
deviation. Correlation between two groups was analyzed using the
Pearson's correlation test. P<0.05 was considered to indicate a
statistically significant difference.
Results
LHX2 expression is increased in NSCLC
cells and tissues
LHX2 is involved in various types of human cancer,
including NSCLC (18). To further
verify whether LHX2 expression is associated with NSCLC, the
present study first examined the expression level of LHX2 in seven
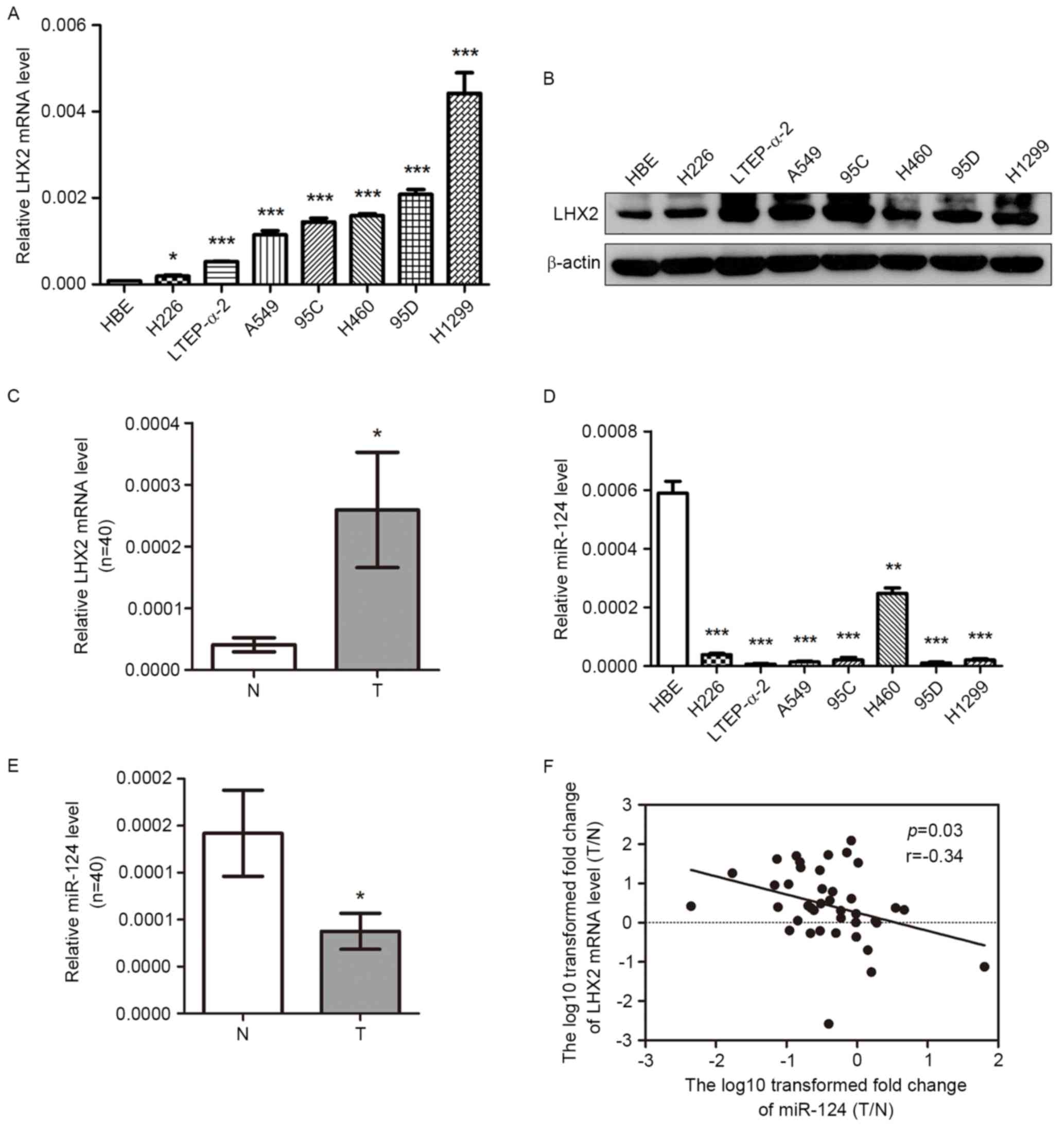
NSCLC cell lines. As shown in Fig. 1A and
B, the levels of LHX2 mRNA and protein expression were
significantly higher in seven NSCLC cell lines (H226, LTEP-a-2,
A549, 95C, H460, 95D and H1299) compared with the control HBE
cells. Subsequently, the level of LHX2 mRNA expression was detected
in 40 paired NSCLC tissues and adjacent noncancerous lung tissues,
and this analysis revealed that LHX2 mRNA expression was
significantly higher in NSCLC tissues compared with the paired
noncancerous lung tissues (Fig. 1C).
When classified by various clinicopathological characteristics,
NSCLC samples did not reveal any difference in the LHX2 mRNA
expression level ratio (T/N) (Table
II).
Expression of miR-124 is decreased and
negatively associated with the level of LHX2 expression in NSCLC
cells and tissues
As presented in Fig.
1D, the expression of miR-124 was significantly lower in seven
NSCLC cell lines (H226, LTEP-a-2, A549, 95C, H460, 95D and H1299)
compared with the expression in HBE cells. Furthermore, miR-124
expression level was significantly lower in NSCLC tissues compared
with the paired noncancerous lung tissues (Fig. 1E). No significant difference in
miR-124expression ratio (T/N) was revealed between NSCLC samples
when classified by various clinicopathological characteristics
(Table II). Of note, the ratio of
miR-124 expression level (T/N) was inversely associated with LHX2
mRNA expression level (T/N) in 40 paired tissues (Fig. 1F). The results, analyzed using
Pearson's correlation test, suggested that there may be a
correlation between miR-124 and LHX2 expression levels in
NSCLCs.
miR-124 suppresses LHX2 expression
level by targeting LHX2 3′-UTR in NSCLC cells
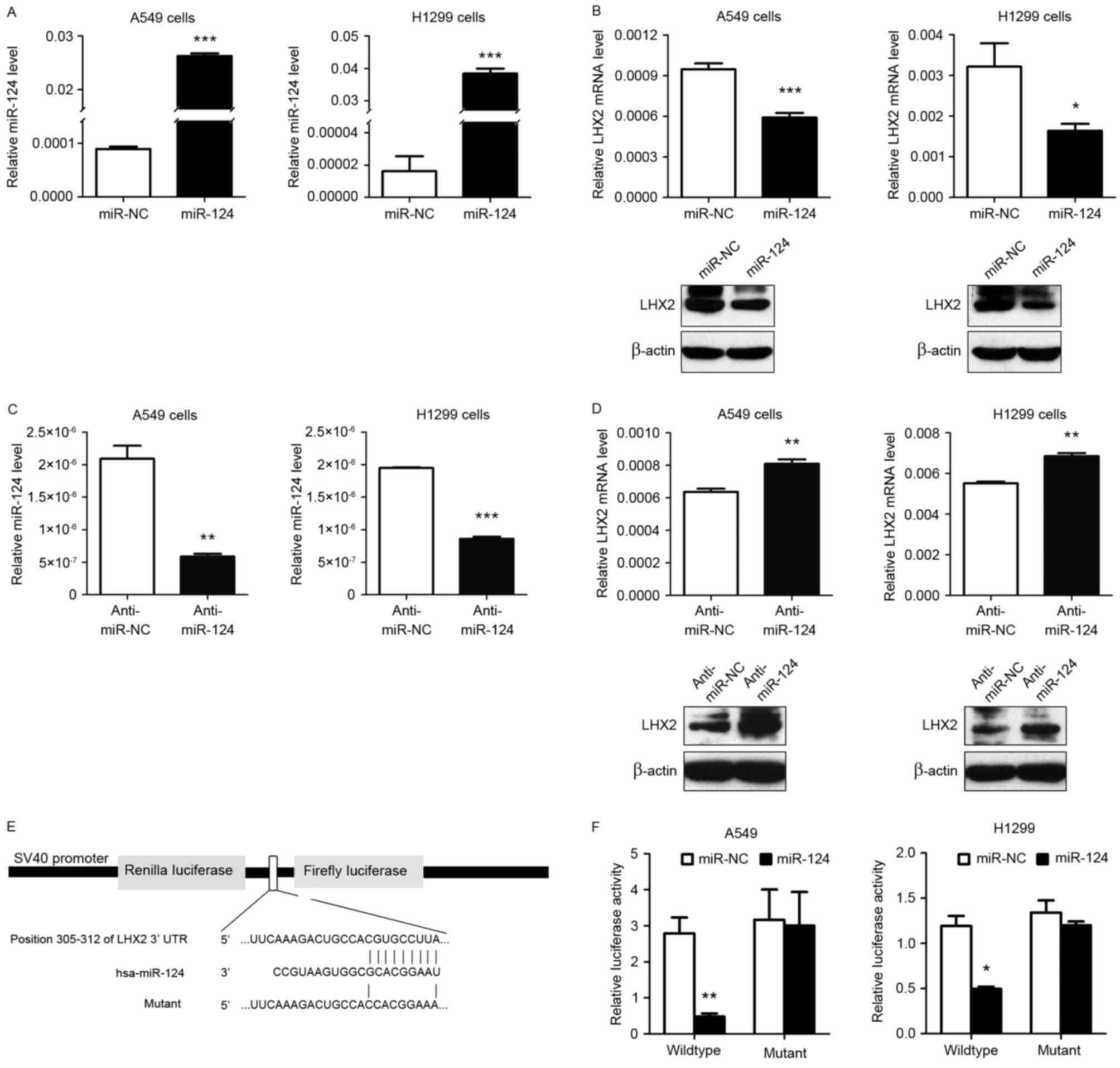
As shown in Fig. 2A, a
markedly increased expression level of miR-124 was observed in A549
and H1299 cells when transiently transfected with miR-124 mimic.
Notably, miR-124 overexpression significantly inhibited LHX2 mRNA
and protein expression levels in A549 and H1299 cells (Fig. 2B), and miR-124 downregulation
(Fig. 2C) markedly promoted LHX2 mRNA
and protein expression levels in A549 and H1299 cells (Fig. 2D), suggesting that miR-124 served an
important role in downregulating LHX2 in NSCLC. To further
investigate the molecular mechanisms underlying this inhibition
effect, the present study first used TargetScanHuman (version 7.0)
software, which identified LHX2 3′-UTR to be a putative target of
miR-124. Therefore, in order to confirm this, the present study
subcloned LHX2 3′-UTR containing the miR-124 binding site
(wild-type/mutant) into psiCHECK-2 vectors (Fig. 2E) and transiently co-transfected the
reporter vector with miR-124 mimic into NSCLC cell lines A549 and
H1299. The results of the luciferase reporter assay demonstrated
that miR-124 significantly inhibited the luciferase activities in
NSCLC cells transfected with the LHX2 3′-UTR wild-type plasmids,
whereas miR-124 did not suppress the luciferase activities in cells
transfected with the mutant vectors (Fig.
2F). Taken together, the results indicated that miR-124 may
directly target the 3′-UTR of LHX2 and thereby reduced the
expression level of LHX2.
miR-124 overexpression or knockdown of
LHX2 represses NSCLC cell migration and invasion
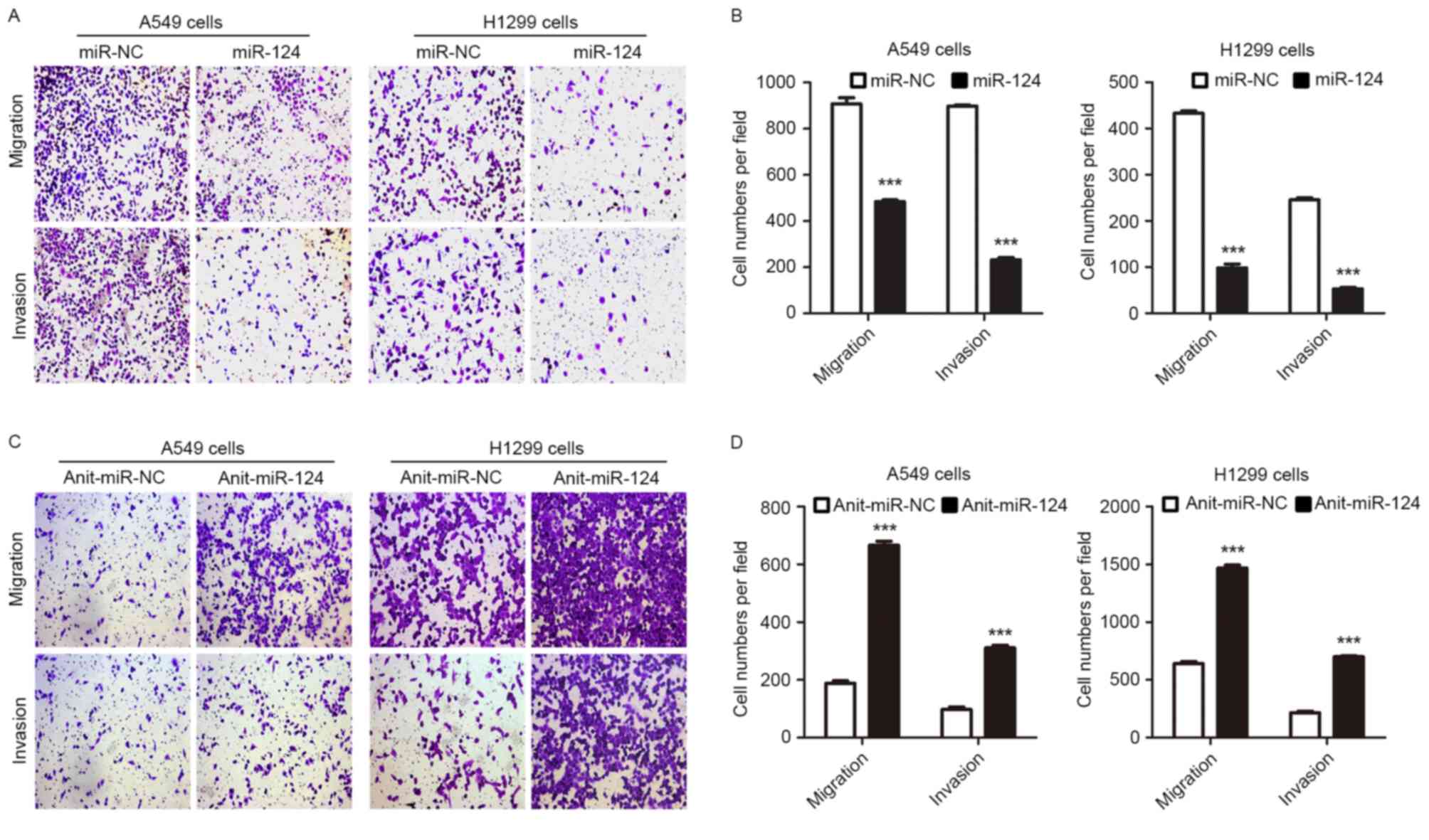
Given the observation that miR-124 may attenuate
NSCLC metastasis by targeting MYO10 (13) and the results of the present study,
whether repression of LHX2 expression by miR-124 may inhibit NSCLC
cell migration and invasion was subsequently investigated. In
support of this hypothesis, the Transwell assay revealed that A549
and H1299 cells overexpressing miR-124 exhibited an impaired
ability to migrate and invade (Fig. 3A
and B). Furthermore, A549 and H1299 cells with downregulated
miR-124 expression demonstrated increased migratory and invasive
abilities (Fig. 3C and D). Notably,
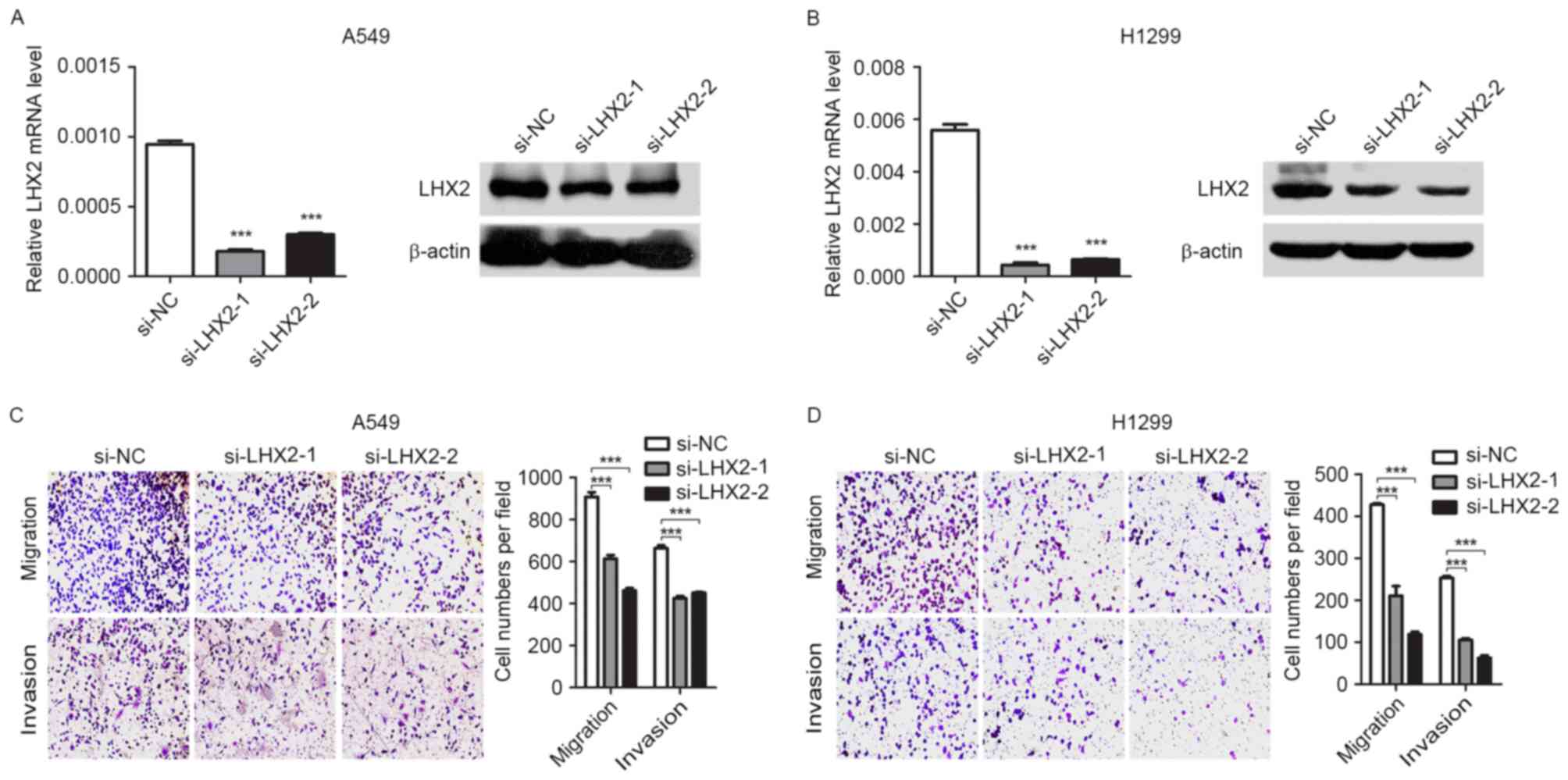
knockdown of LHX2 (Fig. 4A and B)
suppressed migratory and invasive abilities of A549 and H1299 cells
(Fig. 4C and D). Taken together, the
results suggested that miR-124 may attenuate NSCLC cell migration
and invasion by targeting LHX2.
Discussion
LHX2 serves important roles in multiple biological
processes, including embryo development (16), cell fate decision, proliferation
(21) and cell differentiation
(22). Therefore, aberrant expression
of LHX2 may be associated with certain human diseases, including
cancer. High levels of LHX2 were expressed in pancreatic ductal
adenocarcinoma (23). In present
study, it was revealed that LHX2 was highly expressed in NSCLC
cells and tissues. Although the authors of the present study have
previously reported that knockdown of LHX2 attenuated NSCLC cell
proliferation (18), whether LHX2
expression affects NSCLC cell migration and invasion has not yet
been investigated. Kuzmanov et al (17) reported that LHX2 functioned as a
promoter of metastasis in breast cancer cells. In the present
study, the findings indicated that knockdown of LHX2 significantly
inhibited the aggressive abilities of NSCLC cells.
Subsequently, the present study investigated the
miRNA-mediated mechanism underlying LHX2 regulation in NSCLC cell
migration and invasion. Considering the idea that miRNAs serve a
key role in regulating various gene expression levels at the
post-transcriptional level (24) and
in silico prediction which identified the 3′-UTR of LHX2
transcript to be a putative target of miR-124, the present study
performed cell-based and biochemical analyses to confirm this.
Firstly, the present study revealed that LHX2 expression level was
markedly downregulated and upregulated in NSCLC cells when
transiently transfected with miR-124 mimic and miR-124 inhibitor,
respectively. Secondly, luciferase reporter assays demonstrated
that miR-124 repressed LHX2 expression level by targeting a
specific site of the LHX2 3′-UTR. To the best of our knowledge,
this is the first evidence that miR-124 targets LHX2 and inhibits
its expression in NSCLC cells. In addition, the present study
observed that miR-124 overexpression inhibited NSCLC cell migration
and invasion, which was consistent with the results from a previous
study by Sun et al (13). The
phenotype of miR-124 overexpression was the same as the phenotype
of NSCLC cells with knockdown of LHX2, which presented an
attenuated aggressive ability, suggesting that miR-124 may inhibit
NSCLC cell migration and invasion by targeting LHX2. Of note,
miR-124 may also target MYO10 and inhibit NSCLC metastasis
(13). This is not surprising because
single miRNA have been reported to regulate various target genes to
exert its function (25). In further
support of this, miR-124 directly targets ESX/epidermal growth
factor receptor or talin 1 to suppress cell invasion in head and
neck squamous cell carcinoma (11)
and prostate cancer (26).
Recently, a low level of miR-124 expression was
demonstrated to be significantly associated with positive lymph
node metastasis and poor prognosis in human cancer, including NSCLC
(14,15). The present study observed that miR-124
inhibition increased the migratory and invasive abilities of NSCLC
cells, and miR-124 was downregulated in NSCLC tissues. However, due
to the limited sample size, the present study failed to reveal the
significant association of miR-124 expression level with lymph node
metastasis in NSCLC tissues. Furthermore, the results demonstrated
that a low expression level of miR-124 was inversely associated
with a high level of LHX2 expression in NSCLCs.
In conclusion, the present study provided the first
evidence that LHX2 is involved in NSCLC cell migration and
invasion, which was regulated at least partially by
miR-124.Mechanistically, miR-124 reduced LHX2 expression by
directly targeting the LHX2 3′-UTR. The results of the present
study suggest that overexpression of miR-124 or silencing of LHX2
may provide a therapeutic strategy for advanced NSCLC.
Acknowledgements
The present study was supported in part by grants
from the National Natural Science Foundation of China (grant nos.
81372277, 81171894 and81502498), the Jiangsu Province's Key
Provincial Talents Program (grant no. RC2011106), the Science and
Technology Committee of Jiangsu Province (grant no. BK20131159),
the ‘333’ Project of Jiangsu Province Government (grant no.
2011-III-2166), the Graduate Innovation Project of Jiangsu Province
(grant no. CXZZ13_0830), the Natural Science Foundation of the
Jiangsu Higher Education Institution (grant no. 14KJB0017), the
Soochow Scholar Project of Soochow University (grant no.
SSPSU2010-51) and a project funded by the Priority Academic Program
Development of Jiangsu Higher Education Institution
(PAPD-XL2014014).
References
|
1
|
Herbst RS, Heymach JV and Lippman SM: Lung
cancer. N Engl J Med. 359:1367–1380. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
DeSantis CE, Lin CC, Mariotto AB, Siegel
RL, Stein KD, Kramer JL, Alteri R, Robbins AS and Jemal A: Cancer
treatment and survivorship statistics, 2014. CA Cancer J Clin.
64:252–271. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Liu RY, Zeng Y, Lei Z, Wang L, Yang H, Liu
Z, Zhao J and Zhang HT: JAK/STAT3 signaling is required for
TGF-β-induced epithelial-mesenchymal transition in lung cancer
cells. Int J Oncol. 44:1643–1651. 2014.PubMed/NCBI
|
|
4
|
Guo H, Ingolia NT, Weissman JS and Bartel
DP: Mammalian microRNAs predominantly act to decrease target mRNA
levels. Nature. 466:835–840. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wilson RC and Doudna JA: Molecular
mechanisms of RNA interference. Annu Rev Biophys. 42:217–239. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Butz H, Racz K, Hunyady L and Patocs A:
Crosstalk between TGF-β signaling and the microRNA machinery.
Trends Pharmacol Sci. 33:382–393. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lei Z, Xu G, Wang L, Yang H, Liu X, Zhao J
and Zhang HT: MiR-142-3p represses TGF-β-induced growth inhibition
through repression of TGFβR1 in non-small cell lung cancer. FASEB
J. 28:2696–2704. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Arabkheradmand A, Safari A, Seifoleslami
M, Yahaghi E and Gity M: Down-regulated microRNA-124 expression as
predictive biomarker and its prognostic significance with
clinicopathological features in breast cancer patients. Diagn
Pathol. 10:1782015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ibarrola-Villava M, Llorca-Cardeñosa MJ,
Tarazona N, Mongort C, Fleitas T, Perez-Fidalgo JA, Roselló S,
Navarro S, Ribas G and Cervantes A: Deregulation of ARID1A, CDH1,
cMET and PIK3CA and target-related microRNA expression in gastric
cancer. Oncotarget. 6:26935–26945. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wang X, Wu Q, Xu B, Wang P, Fan W, Cai Y,
Gu X and Meng F: miR-124 exerts tumor suppressive functions on the
cell proliferation, motility and angiogenesis of bladder cancer by
fine-tuning UHRF1. FEBS J. 282:4376–4388. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhang M, Piao L, Datta J, Lang JC, Xie X,
Teknos TN, Mapp AK and Pan Q: miR-124 regulates the
epithelial-restricted with serine box/epidermal growth factor
receptor signaling axis in head and neck squamous cell carcinoma.
Mol Cancer Ther. 14:2313–2320. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Xi ZW, Xin SY, Zhou LQ, Yuan HX, Wang Q
and Chen KX: Downregulation of rho-associated protein kinase 1 by
miR-124 in colorectal cancer. World J Gastroenterol. 21:5454–5464.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Sun Y, Ai X, Shen S and Lu S:
NF-κB-mediated miR-124 suppresses metastasis of non-small-cell lung
cancer by targeting MYO10. Oncotarget. 6:8244–8254. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Dong LL, Chen LM, Wang WM and Zhang LM:
Decreased expression of microRNA-124 is an independent unfavorable
prognostic factor for patients with breast cancer. Diagn Pathol.
10:452015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhang Y, Li H, Han J and Zhang Y:
Down-regulation of microRNA-124 is correlated with tumor metastasis
and poor prognosis in patients with lung cancer. Int J Clin Exp
Pathol. 8:1967–1972. 2015.PubMed/NCBI
|
|
16
|
Porter FD, Drago J, Xu Y, Cheema SS,
Wassif C, Huang SP, Lee E, Grinberg A, Massalas JS, Bodine D, et
al: Lhx2, a LIM homeobox gene, is required for eye, forebrain, and
definitive erythrocyte development. Development. 124:2935–2944.
1997.PubMed/NCBI
|
|
17
|
Kuzmanov A, Hopfer U, Marti P,
Meyer-Schaller N, Yilmaz M and Christofori G: LIM-homeobox gene 2
promotes tumor growth and metastasis by inducing autocrine and
paracrine PDGF-B signaling. Mol Oncol. 8:401–416. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Shi X, Zhan L, Xiao C, Lei Z, Yang H, Wang
L, Zhao J and Zhang HT: miR-1238 inhibits cell proliferation by
targeting LHX2 in non-small cell lung cancer. Oncotarget.
6:19043–19054. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Mountain CF: Revisions in the
international system for staging lung cancer. Chest. 111:1710–1717.
1997. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
do Pinto OP, Kolterud A and Carlsson L:
Expression of the LIM-homeobox gene LH2 generates immortalized
steel factor-dependent multipotent hematopoietic precursors. EMBO
J. 17:5744–5756. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Chou SJ and O'Leary DD: Role for Lhx2 in
corticogenesis through regulation of progenitor differentiation.
Mol Cell Neurosci. 56:1–9. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhou F, Gou S, Xiong J, Wu H, Wang C and
Liu T: Oncogenicity of LHX2 in pancreatic ductal adenocarcinoma.
Mol Biol Rep. 41:8163–8167. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ohtsuka M, Ling H, Doki Y, Mori M and
Calin GA: MicroRNA processing and human cancer. J Clin Med.
4:1651–1667. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Lim LP, Lau NC, Garrett-Engele P, Grimson
A, Schelter JM, Castle J, Bartel DP, Linsley PS and Johnson JM:
Microarray analysis shows that some microRNAs downregulate large
numbers of target mRNAs. Nature. 433:769–773. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhang W, Mao YQ, Wang H, Yin WJ, Zhu SX
and Wang WC: MiR-124 suppresses cell motility and adhesion by
targeting talin 1 in prostate cancer cells. Cancer Cell Int.
15:492015. View Article : Google Scholar : PubMed/NCBI
|