Introduction
Intervertebral disc (IVD) disease is a common
surgical disease, and IVD degeneration is hypothesized to be the
first step in degenerative spinal changes (1). IVD degeneration is typically associated
with disc herniation and back pain, which has a marked effect on
the sufferer's life and causes severe chronic pain (2). Furthermore, lower back pain may limit
the activity of individuals <45 years, which has a marked
socio-economic impact (3). The
etiology of lower back pain is unclear, but in 40% of cases it is
associated with IVD degeneration (4).
It is reported that ~40% of individuals <30 years of age, and
>90% of individuals >55 years, exhibit moderate-to-severe
levels of IVD degeneration (5,6). It is
estimated that the costs associated with lumbar disc and lower back
disorders exceed $100 billion/year in the USA (7). Traditionally, inflammation has mostly
been observed as detrimental and is associated with disease
progression. Although the etiology remains unknown, the osmolality
has been identified to be associated with IVD disease (8–10). Katz
(7) reported that the osmotic
environment exerted an appreciable effect on gene expression and
also affected responses to mechanical stimuli. Mavrogonatou and
Kletsas (11) indicated that high
osmolality was able to activate the G1 and G2
cell cycle checkpoints and affect the DNA integrity of nucleus
pulposus IVD cells, triggering an enhanced DNA repair response.
Although changes in the extracellular osmolality markedly
influenced the behavior of IVD cells, their response to this
condition has not yet been fully elucidated. In the present study,
the associated biological processes and potential biomarkers in the
response of IVD to osmotic stimuli were investigated using gene
expression analysis.
Materials and methods
Gene expression profile
The gene expression dataset GSE1648 (8) was downloaded from the Gene Expression
Omnibus (http://www.ncbi.nlm.nih.gov/geo/) database. There was
a total of 11 IVD cell samples in GSE1648, including 4 hyperosmotic
stimuli samples, 3 hypoosmotic stimuli samples and 4 isosmotic
stimuli samples.
Data pre-processing and identification
of differentially expressed genes (DEGs)
For the expression profile, the original data were
converted into a recognizable format using R software version 3.1.1
(from bioconductor.org/packages/release/bioc/html/biomaRt.html),
and the Affy package (12) version
1.48.0 (from
bioconductor.org/packages/release/bioc/html/affy.html) was used
for the background correction and normalization. The DEGs in
hyperosmotic (designated DEGs-hyper) or hypoosmotic (DEGs-hypo) IVD
cells, compared with isosmotic cells, were identified using the
limma package (13) version 3.18.13
(from bioconductor.org/packages/release/bioc/html/limma.html)
based on gene expression differences of P<0.05 and
|log2(fold-change)|>0.05.
Gene Ontology (GO) term enrichment
analysis
Functional annotation of DEGs is a necessary and
critical step in the analysis of microarray data. The Database for
Annotation, Visualization and Integrated Discovery (DAVID,
http://david.abcc.ncifcrf.gov/)
(14) is a tool providing a
comprehensive set of functional annotation. Enriched GO terms from
DEGs-hyper were identified using DAVID with a threshold of
P<0.05.
Construction of the protein-protein
interaction (PPI) network
The Human Protein Reference Database (HPRD) was used
as a centralized platform to visually depict and integrate
information pertaining to domain architecture, post-translational
modifications, interaction networks and disease association for
each protein in the human proteome (15). PPI network data for DEGs-hyper was
obtained from HPRD and visualized using Cytoscape (16) software.
Establishment and analysis of the
microRNA (miRNA)-regulated-gene network
The software TargetScan (17) was used to predict biological targets
of miRNAs by searching for the presence of conserved 8-mer and
7-mer sites that matched the seed region of each miRNA. TargetScan
screened for miRNAs associated with the regulation of DEGs-hyper,
and miRNA-gene connections were obtained. An miRNA-regulated-gene
network was established and visualized using Cytoscape software,
and every node was analyzed according to the number of connections
with other nodes (degree). miRNAs for which the degree was ≥1 were
removed from the analysis.
Results
DEGs
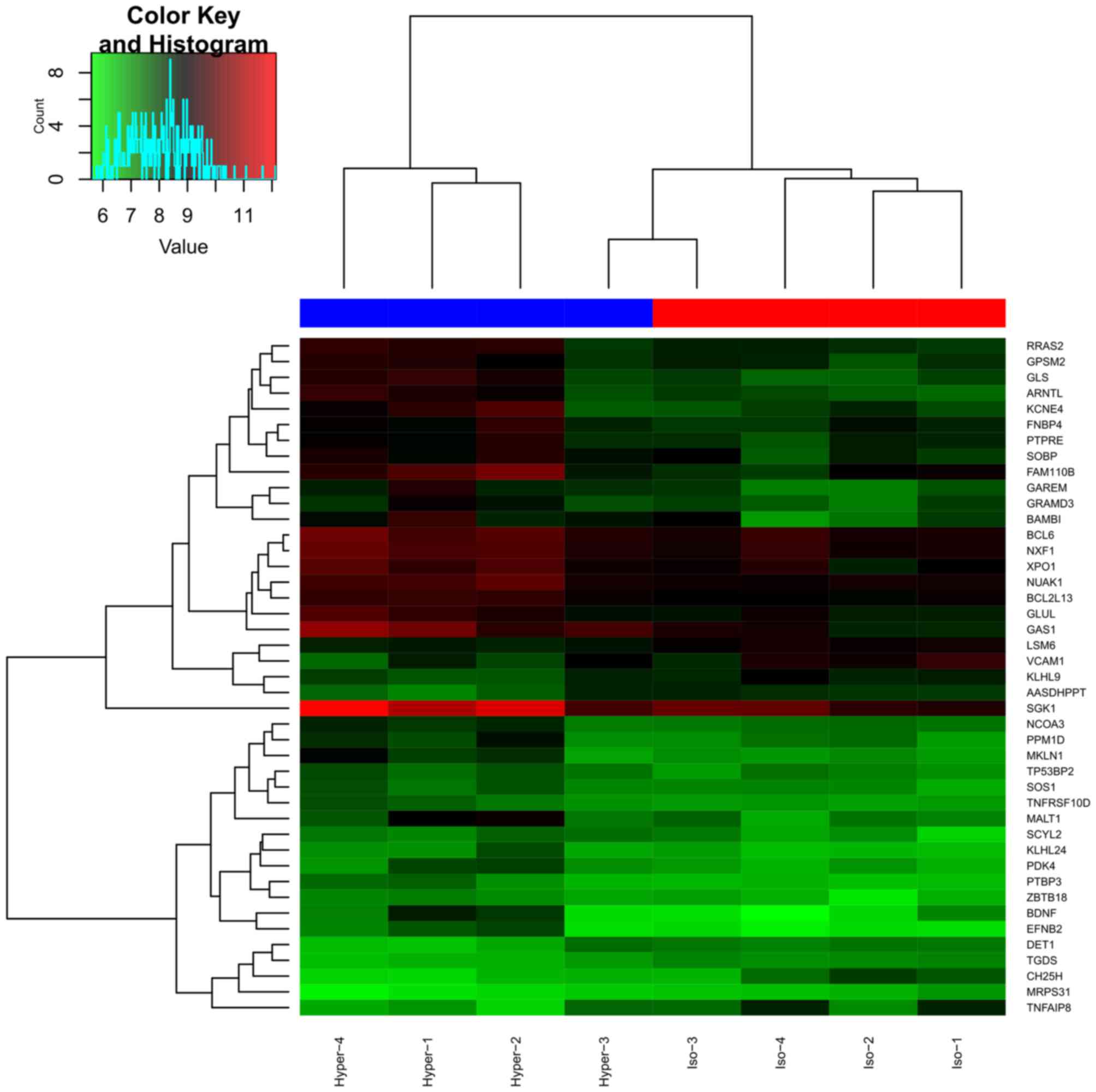
A total of 43 DEGs (34 up- and 9 downregulated) were
identified in DEGs-hyper, and the cluster graph is presented in
Fig. 1. However, only 9 DEGs were
obtained in DEGs-hypo, which were not studied further.
GO terms of DEGs-hyper
In total, 41 GO terms were enriched in DEGs-hyper,
and the 10 most significantly enriched GO terms (e.g., regulation
of apoptosis, regulation of programmed cell death and regulation of
cell death) are presented in Table
I.
 | Table I.Top 10 most significantly enriched GO
terms of differentially expressed genes in intervertebral disc
cells of hyperosmotic stimuli samples compared with the isosmotic
stimuli. |
Table I.
Top 10 most significantly enriched GO
terms of differentially expressed genes in intervertebral disc
cells of hyperosmotic stimuli samples compared with the isosmotic
stimuli.
| Category | GO ID | GO name | Number of
genes | P-value |
|---|
| BP | GO:0042981 | Regulation of
apoptosis | 9 |
3.48×10−4 |
| BP | GO:0043067 | Regulation of
programmed cell death | 9 |
3.73×10−4 |
| BP | GO:0010941 | Regulation of cell
death | 9 |
3.82×10−4 |
| BP | GO:0006913 | Nucleocytoplasmic
transport | 5 |
4.20×10−4 |
| BP | GO:0051169 | Nuclear
transport | 5 |
4.41×10−4 |
| BP | GO:0012501 | Programmed cell
death | 7 | 0.00232 |
| BP | GO:0008219 | Cell death | 7 | 0.00519 |
| BP | GO:0030183 | B cell
differentiation | 3 | 0.00537 |
| BP | GO:0016265 | Death | 7 | 0.00537 |
| BP | GO:0051168 | Nuclear export | 3 | 0.00828 |
PPI network of DEGs-hyper
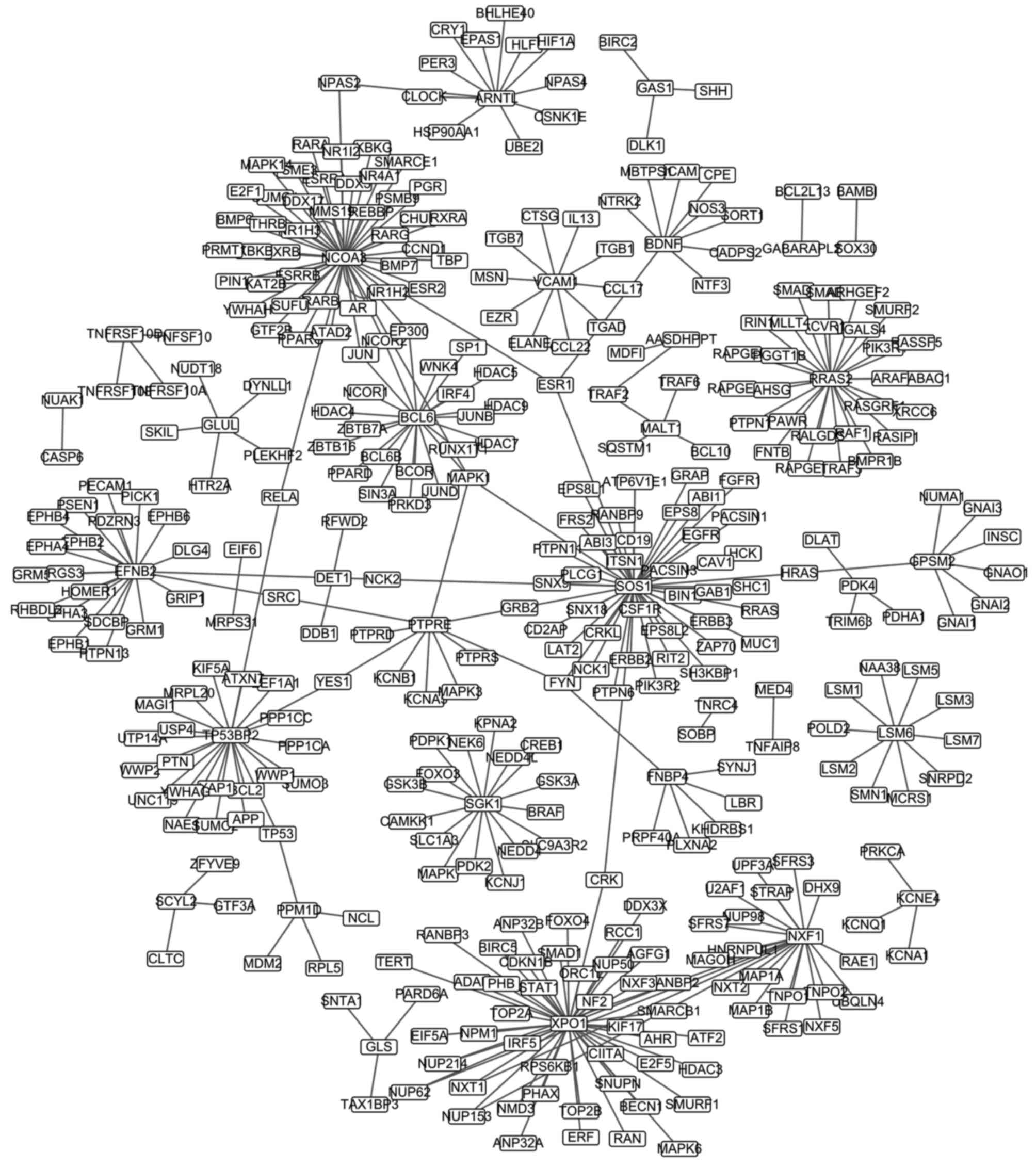
The PPI network was constructed and is presented in
Fig. 2, and included 376 pairs and
382 nodes. The top 20 nodes (e.g., NCOA3, SOS1 and
XPO1), as defined by highest degree, are presented in
Table II.
 | Table II.Top 20 nodes with a higher degree in
the protein-protein interaction network. |
Table II.
Top 20 nodes with a higher degree in
the protein-protein interaction network.
| Gene | Degree |
|---|
| NCOA3 | 49 |
| SOS1 | 45 |
| XPO1 | 45 |
| RRAS2 | 27 |
| TP53BP2 | 23 |
| BCL6 | 22 |
| NXF1 | 22 |
| EFNB2 | 21 |
| SGK1 | 16 |
| ARNTL | 13 |
| LSM6 | 10 |
| PTPRE | 10 |
| VCAM1 | 10 |
| BDNF | 9 |
| GPSM2 | 7 |
| FNBP4 | 6 |
| GLUL | 5 |
| MALT1 | 4 |
| MAPK1 | 4 |
| PPM1D | 4 |
miRNA-regulated-gene network of
DEGs-hyper
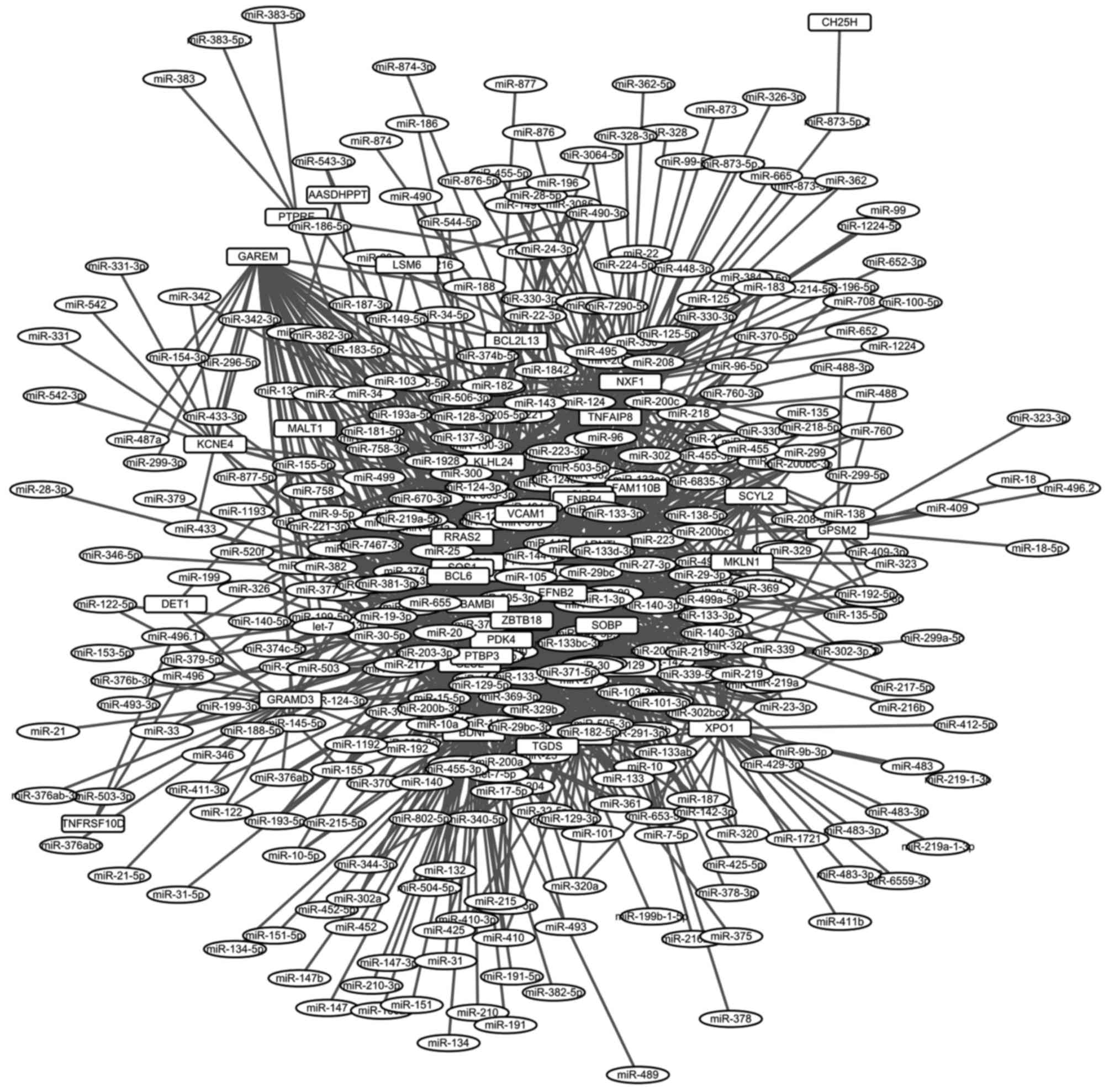
In total, 1,314 miRNA-regulated-gene connections
were identified with TargetScan. A miRNA-gene-regulated network was
established, including 1,314 connections and 422 nodes (Fig. 3). The top 20 nodes (e.g., ZBTB18,
EFNB2 and SOBP) in the regulated network, as determined
by degree, are presented in Table
III.
 | Table III.Top 20 nodes with higher degree in
the microRNA-gene-regulated network. |
Table III.
Top 20 nodes with higher degree in
the microRNA-gene-regulated network.
| Gene | Degree |
|---|
| ZBTB18 | 81 |
| EFNB2 | 73 |
| SOBP | 72 |
| NXF1 | 68 |
| BDNF | 66 |
| PTBP3 | 64 |
| PPM1D | 59 |
| BAMBI | 58 |
| NCOA3 | 55 |
| KLHL24 | 47 |
| SGK1 | 45 |
| GAREM | 42 |
| NUAK1 | 42 |
| SOS1 | 42 |
| BCL6 | 38 |
| XPO1 | 37 |
| FAM110B | 34 |
| TNFAIP8 | 34 |
| KLHL9 | 29 |
Discussion
The normal upper limit of water in the human IVD is
~500 mOsm/kg, which is higher than that routinely encountered in
the majority of other parts of the body (~300 mOsm/kg) (18). It was previously reported that ~18% of
the fluid of IVD was lost and re-imbibed during a diurnal cycle,
with consequent changes in osmolality (19). Changes in osmolality are an important
component of the physicochemical environment of IVD, as variations
in disc loading leads to alterations of disc hydration. A study of
IVD cells in a three-dimensional alginate culture system confirmed
that the biological response to altered osmolarity is mediated, in
part, by changes at the transcriptional level (20). In the present study, 43 DEGs were
identified in hyperosmotic cells, whereas 9 DEGs were identified in
hypoosmotic cells, compared with isosmotic cells. These results
suggest that IVD is more sensitive to hyperosmotic stimuli than to
hypoosmotic stimuli, and that the effects of hyperosmotic stimuli
may be far greater at the transcriptional level. Hyperosmotic
stimuli were previously demonstrated to elicit calcium transience
in IVD cells that were modulated by the stability of the actin
cytoskeleton (21,22). In the present study, DEGs-hyper were
the basis of further research, whereas DEGs-hypo were not further
studied.
The enriched GO terms for DEGs-hyper were
predominantly associated with the biological processes of apoptosis
and cell death, and the regulation of these processes. Three
subfamilies were identified: Extracellular-signal-regulated kinase
(ERK1/2), p38 mitogen-activated protein kinase (p38 MAPK, p38) and
c-Jun N-terminal kinase (JNK1/2). Nucleus pulposus cell apoptosis
is one of the changes associated with IVD degeneration (23). p38 and JNK1/2 are serine/threonine
protein kinases activated by various stress stimuli, including
osmotic shock, toxic compounds and pro-inflammatory cytokines
(24). ERK1/2 was previously
demonstrated to be activated by high osmolality in nucleus pulposus
cells in vitro (25). Dong
et al (26) reported that high
osmolality activated p38 MAPK, JNK1/2 and ERK1/2 in rabbit nucleus
pulposus cells. The activated p38 MAPK and JNK1/2 induced cell
apoptosis; by contrast, the activation of ERK1/2 promoted cell
survival. A recent study (27)
indicated that the effects of osmolality on nucleus pulposus cell
apoptosis depended on the osmolality level (hypo-, iso- or hyper-)
and osmolality mode (constant or cyclic). Furthermore, inhibition
of the ERK1/2 pathway promoted nucleus pulposus cell apoptosis in
this process (27). Therefore, it was
suspected that the biological processes of apoptosis and cell death
may be associated with the effect of hyperosmolality on IVD
diseases.
The PPI network was analyzed, and nuclear receptor
coactivator 3 (NCOA3), SOS Ras/Rac guanine nucleotide exchange
factor 1 (SOS1) and exportin 1 (XPO1) were the top three nodes as
defined by degree. NCOA3 was identified to be associated with the
pathophysiology of osteoarthritis (OA) (28,29). A
recent study concluded that NCOA3 was subject to a
cis-acting expression quantitative trait locus in articular
cartilage, which was associated with the OA association signal and
associated with the OA-associated allele of the functional
single-nucleotide polymorphism rs116855380 (30). Although there is no direct evidence
that NCOA3 is associated with IVD degeneration, IVD degeneration
and OA may exhibit similarities in their occurrence and
development, as they are joint degeneration diseases. SOS1
primarily encoded membrane-bound guanine nucleotide-binding
proteins in humans, which function in the transduction of signals
that control cell growth and differentiation (31).
It was previously reported that SOS1 participates in
the process of apoptosis (32,33). It
was identified that the biological processes of apoptosis and cell
death may be associated with the effect of high osmolality on IVD
diseases. The mutation of SOS1 serves an important role in
the salt-tolerance and osmotic stimuli of Arabidopsis and
tobacco (34–37). XPO1 is a specific receptor for
leucine-rich nuclear export sequences (38) and was identified to mediate the
nuclear export of proteins in a range of species (39,40).
Ferrigno et al (41) indicated
that XPO1 mediated the export of high osmolality glycerol response
MAPK to the cytoplasm of cells adapted to hyper-osmotic stimuli.
Thus, NCOA3, SOS1 and XPO1 may be genes
directly affected by osmotic stimuli in IVD cells, although more
trials and clinical validation are required. Similarly,
ZBTB18, EFNB2 and SOBP were the top 3 nodes in
the miRNA-gene-regulated network, and they may exhibit an intimate
association with the effects of osmotic stimuli on IVD.
The dataset GSE1648 was created by Boyd et al
(8); however, the present study
adopted different methods and obtained novel results compared with
the Boyd et al study (8).
First, differential expression analysis was performed for cells in
hyperosmotic and hypoosmotic conditions in the present study,
whereas Boyd et al (8) only
conducted differential expression analysis of cells in hyperosmotic
conditions. Secondly, the PPI network and miRNA-gene-regulated
network were constructed in the present study; however, this was
not carried out in the study by Boyd et al (8). Finally, potential target genes
(NCOA3, SOS1, XPO1, ZBTB18,
EFNB2 and SOBP) were identified from the PPI network
and the miRNA-gene-regulated network, whereas they were not
identified in the study by Boyd et al (8). These genes exhibited more interactions
compared with those identified by Boyd et al, and therefore
they were considered more reliable.
References
|
1
|
Boos N, Weissbach S, Rohrbach H, Weiler C,
Spratt KF and Nerlich AG: Classification of age-related changes in
lumbar intervertebral discs: 2002 Volvo Award in basic science.
Spine (Phila Pa 1976). 27:2631–2644. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Stewart WF, Ricci JA, Chee E, Morganstein
D and Lipton R: Lost productive time and cost due to common pain
conditions in the US workforce. JAMA. 290:2443–2454. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Andersson GB: Epidemiological features of
chronic low-back pain. Lancet. 354:581–585. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Schwarzer AC, Aprill CN, Derby R, Fortin
J, Kine G and Bogduk N: The prevalence and clinical features of
internal disc disruption in patients with chronic low back pain.
Spine (Phila Pa 1976). 20:1878–1883. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Stolworthy DK, Bowden AE, Roeder BL,
Robinson TF, Holland JG, Christensen SL, Beatty AM, Bridgewater LC,
Eggett DL, Wendel JD, et al: MRI evaluation of spontaneous
intervertebral disc degeneration in the alpaca cervical spine. J
Orthop Res. 33:1776–1783. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Cheung KM, Karppinen J, Chan D, Ho DW,
Song YQ, Sham P, Cheah KS, Leong JC and Luk KD: Prevalence and
pattern of lumbar magnetic resonance imaging changes in a
population study of one thousand forty-three individuals. Spine
(Phila Pa 1976). 34:934–940. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Katz JN: Lumbar disc disorders and
low-back pain: Socioeconomic factors and consequences. J Bone Joint
Surg Am. 88:(Suppl 2). S21–S24. 2006. View Article : Google Scholar
|
|
8
|
Boyd LM, Richardson WJ, Chen J, Kraus VB,
Tewari A and Setton LA: Osmolarity regulates gene expression in
intervertebral disc cells determined by gene array and real-time
quantitative RT-PCR. Ann Biomed Eng. 33:1071–1077. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Setton LA and Chen J: Mechanobiology of
the intervertebral disc and relevance to disc degeneration. J Bone
Joint Surg Am. 88:(Suppl 2). S52–S57. 2006. View Article : Google Scholar
|
|
10
|
Ma H, Xie YZ, Zhao J and Ye B: Small
molecule-enrichment analysis in response to osmotic stimuli in the
intervertebral disc. Genet Mol Res. 11:3668–3675. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Mavrogonatou E and Kletsas D: High
osmolality activates the G1 and G2 cell cycle checkpoints and
affects the DNA integrity of nucleus pulposus intervertebral disc
cells triggering an enhanced DNA repair response. DNA Repair
(Amst). 8:930–943. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Gautier L, Cope L, Bolstad BM and Irizarry
RA: affy-analysis of Affymetrix GeneChip data at the probe level.
Bioinformatics. 20:307–315. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Diboun I, Wernisch L, Orengo CA and
Koltzenburg M: Microarray analysis after RNA amplification can
detect pronounced differences in gene expression using limma. BMC
Genomics. 7:2522006. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Sherman BT, da Huang W, Tan Q, Guo Y, Bour
S, Liu D, Stephens R, Baseler MW, Lane HC and Lempicki RA: DAVID
Knowledgebase: A gene-centered database integrating heterogeneous
gene annotation resources to facilitate high-throughput gene
functional analysis. BMC Bioinformatics. 8:4262007. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Prasad Keshava TS, Goel R, Kandasamy K,
Keerthikumar S, Kumar S, Mathivanan S, Telikicherla D, Raju R,
Shafreen B, Venugopal A, et al: Human protein reference
database-2009 update. Nucleic Acids Res. 37:(Database issue).
D767–D772. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lewis BP, Shih IH, Jones-Rhoades MW,
Bartel DP and Burge CB: Prediction of mammalian microRNA targets.
Cell. 115:787–798. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Urban JP: The role of the physicochemical
environment in determining disc cell behaviour. Biochem Soc Trans.
30:858–864. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
McMillan DW, Garbutt G and Adams MA:
Effect of sustained loading on the water content of intervertebral
discs: Implications for disc metabolism. Ann Rheum Dis. 55:880–887.
1996. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Chen J, Baer AE, Paik PY, Yan W and Setton
LA: Matrix protein gene expression in intervertebral disc cells
subjected to altered osmolarity. Biochem Biophys Res Commun.
293:932–938. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Pritchard S, Erickson GR and Guilak F:
Hyperosmotically induced volume change and calcium signaling in
intervertebral disk cells: The role of the actin cytoskeleton.
Biophys J. 83:2502–2510. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Pritchard S and Guilak F: The role of
F-actin in hypo-osmotically induced cell volume change and calcium
signaling in anulus fibrosus cells. Ann Biomed Eng. 32:103–111.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Freemont AJ: The cellular pathobiology of
the degenerate intervertebral disc and discogenic back pain.
Rheumatology (Oxford). 48:5–10. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Johnson NL, Gardner AM, Diener KM,
Lange-Carter CA, Gleavy J, Jarpe MB, Minden A, Karin M, Zon LI and
Johnson GL: Signal transduction pathways regulated by
mitogen-activated/extracellular response kinase kinase kinase
induce cell death. J Biol Chem. 271:3229–3237. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Hiyama A, Gogate SS, Gajghate S, Mochida
J, Shapiro IM and Risbud MV: BMP-2 and TGF-beta stimulate
expression of beta1,3-glucuronosyl transferase 1 (GlcAT-1) in
nucleus pulposus cells through AP1, TonEBP and Sp1: Role of MAPKs.
J Bone Miner Res. 25:1179–1190. 2010.PubMed/NCBI
|
|
26
|
Dong ZH, Wang DC, Liu TT, Li FH, Liu RL,
Wei JW and Zhou CL: The roles of MAPKs in rabbit nucleus pulposus
cell apoptosis induced by high osmolality. Eur Rev Med Pharmacol
Sci. 18:2835–2845. 2014.PubMed/NCBI
|
|
27
|
Li P, Gan Y, Wang H, Xu Y, Li S, Song L,
Zhang C, Ou Y, Wang L and Zhou Q: Role of the ERK1/2 pathway in
osmolarity effects on nucleus pulposus cell apoptosis in a disc
perfusion culture. J Orthop Res. 35:86–92. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Evangelou E, Kerkhof HJ, Styrkarsdottir U,
Ntzani EE, Bos SD, Esko T, Evans DS, Metrustry S, Panoutsopoulou K,
Ramos YF, et al: A meta-analysis of genome-wide association studies
identifies novel variants associated with osteoarthritis of the
hip. Ann Rheum Dis. 73:2130–2136. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Tsezou A: Osteoarthritis year in review
2014: Genetics and genomics. Osteoarthritis Cartilage.
22:2017–2024. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Gee F, Rushton MD, Loughlin J and Reynard
LN: Correlation of the osteoarthritis susceptibility variants that
map to chromosome 20q13 with an expression quantitative trait locus
operating on NCOA3 and with functional variation at the
polymorphism rs116855380. Arthritis Rheumatol. 67:2923–2932. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Pierre S, Bats AS and Coumoul X:
Understanding SOS (Son of Sevenless). Biochem Pharmacol.
82:1049–1056. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Xiao Z, Li L, Li Y, Zhou W, Cheng J, Liu
F, Zheng P, Zhang Y and Che Y: Rasfonin, a novel 2-pyrone
derivative, induces ras-mutated Panc-1 pancreatic tumor cell death
in nude mice. Cell Death Dis. 5:e12412014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Chang YL, Ho BC, Sher S, Yu SL and Yang
PC: miR-146a and miR-370 coordinate enterovirus 71-induced cell
apoptosis through targeting SOS1 and GADD45beta. Cell Microbiol.
17:802–818. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Yue Y, Zhang M, Zhang J, Duan L and Li Z:
SOS1 gene overexpression increased salt tolerance in transgenic
tobacco by maintaining a higher K(+)/Na(+) ratio. J Plant Physiol.
169:255–261. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Ariga H, Katori T, Yoshihara R, Hase Y,
Nozawa S, Narumi I, Iuchi S, Kobayashi M, Tezuka K, Sakata Y, et
al: Arabidopsis sos1 mutant in a salt-tolerant accession revealed
an importance of salt acclimation ability in plant salt tolerance.
Plant Signal Behav. 8:e247792013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Kim JH, Nguyen NH, Jeong CY, Nguyen NT,
Hong SW and Lee H: Loss of the R2R3 MYB, AtMyb73, causes
hyper-induction of the SOS1 and SOS3 genes in response to high
salinity in Arabidopsis. J Plant Physiol. 170:1461–1465. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Katschnig D, Bliek T, Rozema J and Schat
H: Constitutive high-level SOS1 expression and absence of HKT1;1
expression in the salt-accumulating halophyte Salicornia
dolichostachya. Plant Sci. 234:144–154. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Stade K, Ford CS, Guthrie C and Weis K:
Exportin 1 (Crm1p) is an essential nuclear export factor. Cell.
90:1041–1050. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Engel K, Kotlyarov A and Gaestel M:
Leptomycin B-sensitive nuclear export of MAPKAP kinase 2 is
regulated by phosphorylation. EMBO J. 17:3363–3371. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Toone WM, Kuge S, Samuels M, Morgan BA,
Toda T and Jones N: Regulation of the fission yeast transcription
factor Pap1 by oxidative stress: Requirement for the nuclear export
factor Crm1 (Exportin) and the stress-activated MAP kinase
Sty1/Spc1. Genes Dev. 12:1453–1463. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Ferrigno P, Posas F, Koepp D, Saito H and
Silver PA: Regulated nucleo/cytoplasmic exchange of HOG1 MAPK
requires the importin beta homologs NMD5 and XPO1. EMBO J.
17:5606–5614. 1998. View Article : Google Scholar : PubMed/NCBI
|