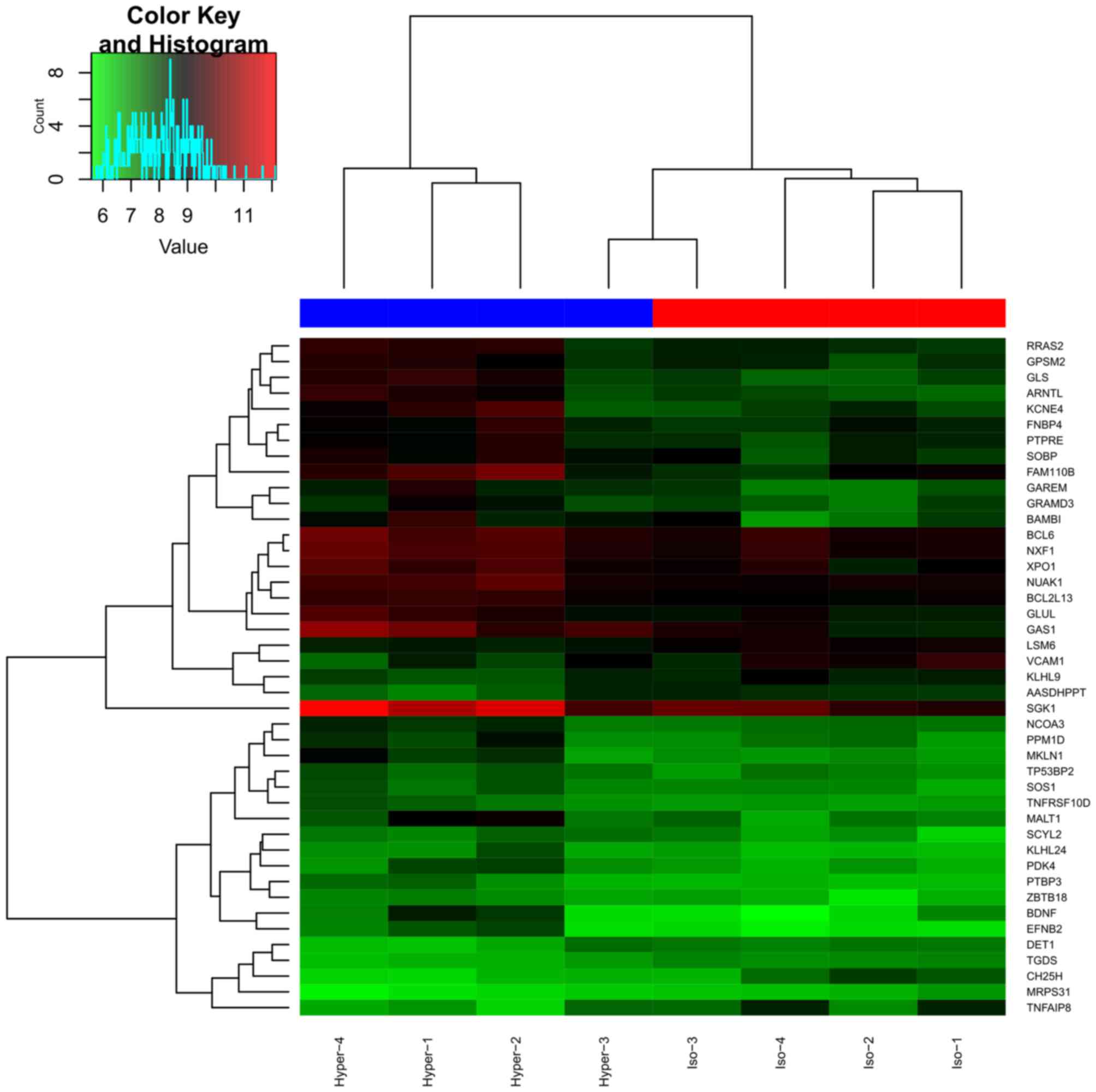
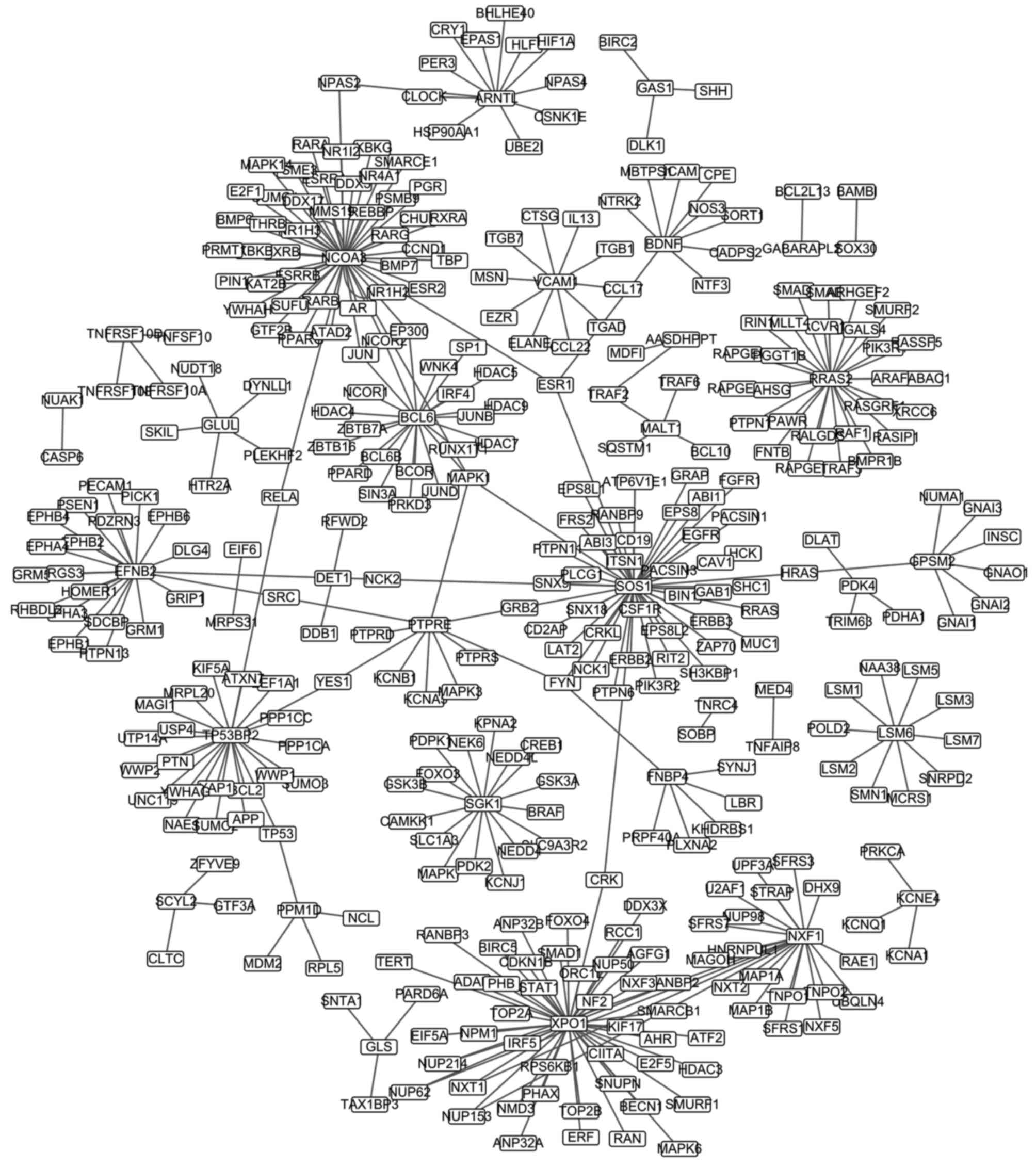
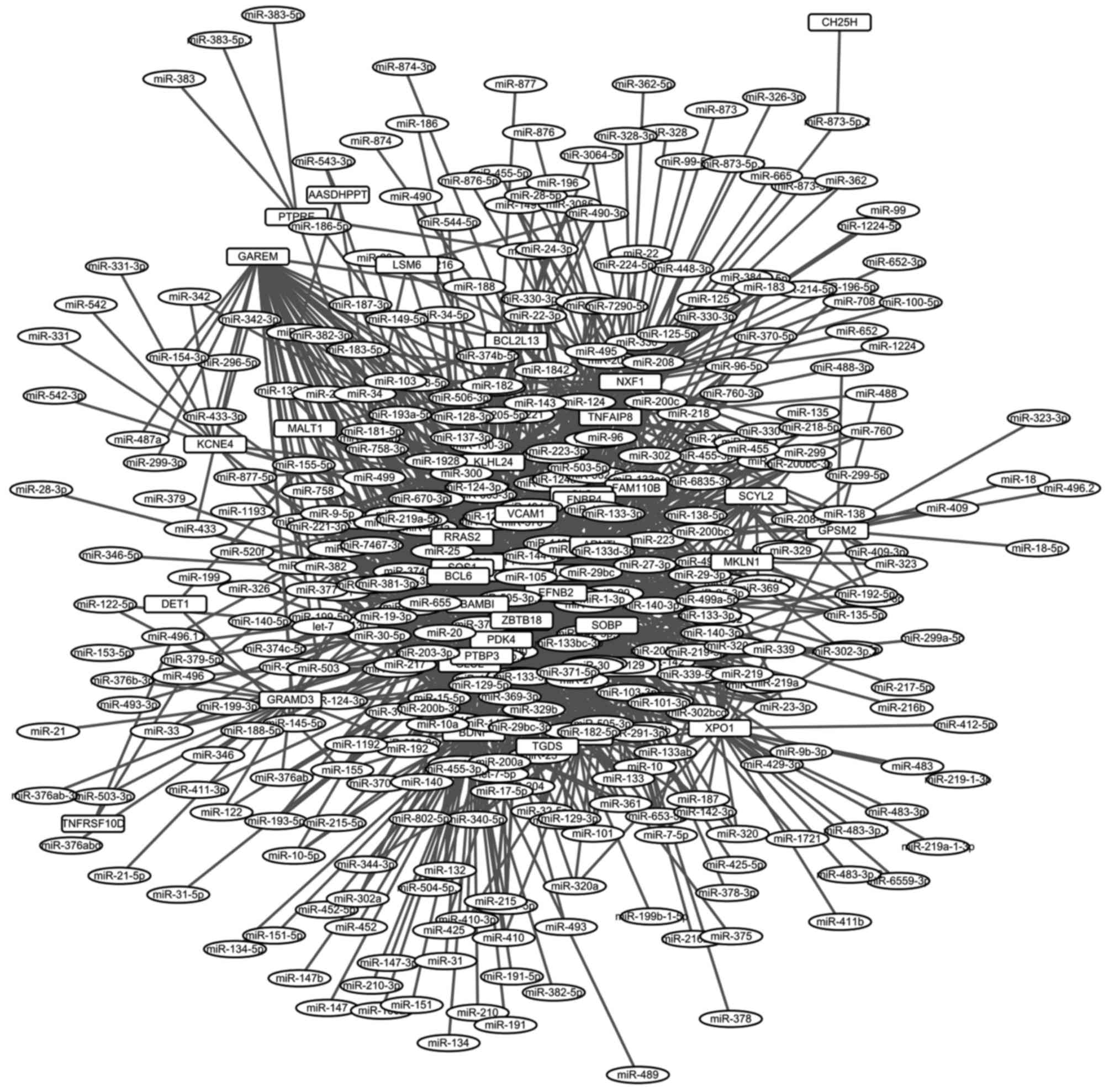
|
1
|
Boos N, Weissbach S, Rohrbach H, Weiler C,
Spratt KF and Nerlich AG: Classification of age-related changes in
lumbar intervertebral discs: 2002 Volvo Award in basic science.
Spine (Phila Pa 1976). 27:2631–2644. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Stewart WF, Ricci JA, Chee E, Morganstein
D and Lipton R: Lost productive time and cost due to common pain
conditions in the US workforce. JAMA. 290:2443–2454. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Andersson GB: Epidemiological features of
chronic low-back pain. Lancet. 354:581–585. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Schwarzer AC, Aprill CN, Derby R, Fortin
J, Kine G and Bogduk N: The prevalence and clinical features of
internal disc disruption in patients with chronic low back pain.
Spine (Phila Pa 1976). 20:1878–1883. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Stolworthy DK, Bowden AE, Roeder BL,
Robinson TF, Holland JG, Christensen SL, Beatty AM, Bridgewater LC,
Eggett DL, Wendel JD, et al: MRI evaluation of spontaneous
intervertebral disc degeneration in the alpaca cervical spine. J
Orthop Res. 33:1776–1783. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Cheung KM, Karppinen J, Chan D, Ho DW,
Song YQ, Sham P, Cheah KS, Leong JC and Luk KD: Prevalence and
pattern of lumbar magnetic resonance imaging changes in a
population study of one thousand forty-three individuals. Spine
(Phila Pa 1976). 34:934–940. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Katz JN: Lumbar disc disorders and
low-back pain: Socioeconomic factors and consequences. J Bone Joint
Surg Am. 88:(Suppl 2). S21–S24. 2006. View Article : Google Scholar
|
|
8
|
Boyd LM, Richardson WJ, Chen J, Kraus VB,
Tewari A and Setton LA: Osmolarity regulates gene expression in
intervertebral disc cells determined by gene array and real-time
quantitative RT-PCR. Ann Biomed Eng. 33:1071–1077. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Setton LA and Chen J: Mechanobiology of
the intervertebral disc and relevance to disc degeneration. J Bone
Joint Surg Am. 88:(Suppl 2). S52–S57. 2006. View Article : Google Scholar
|
|
10
|
Ma H, Xie YZ, Zhao J and Ye B: Small
molecule-enrichment analysis in response to osmotic stimuli in the
intervertebral disc. Genet Mol Res. 11:3668–3675. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Mavrogonatou E and Kletsas D: High
osmolality activates the G1 and G2 cell cycle checkpoints and
affects the DNA integrity of nucleus pulposus intervertebral disc
cells triggering an enhanced DNA repair response. DNA Repair
(Amst). 8:930–943. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Gautier L, Cope L, Bolstad BM and Irizarry
RA: affy-analysis of Affymetrix GeneChip data at the probe level.
Bioinformatics. 20:307–315. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Diboun I, Wernisch L, Orengo CA and
Koltzenburg M: Microarray analysis after RNA amplification can
detect pronounced differences in gene expression using limma. BMC
Genomics. 7:2522006. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Sherman BT, da Huang W, Tan Q, Guo Y, Bour
S, Liu D, Stephens R, Baseler MW, Lane HC and Lempicki RA: DAVID
Knowledgebase: A gene-centered database integrating heterogeneous
gene annotation resources to facilitate high-throughput gene
functional analysis. BMC Bioinformatics. 8:4262007. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Prasad Keshava TS, Goel R, Kandasamy K,
Keerthikumar S, Kumar S, Mathivanan S, Telikicherla D, Raju R,
Shafreen B, Venugopal A, et al: Human protein reference
database-2009 update. Nucleic Acids Res. 37:(Database issue).
D767–D772. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lewis BP, Shih IH, Jones-Rhoades MW,
Bartel DP and Burge CB: Prediction of mammalian microRNA targets.
Cell. 115:787–798. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Urban JP: The role of the physicochemical
environment in determining disc cell behaviour. Biochem Soc Trans.
30:858–864. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
McMillan DW, Garbutt G and Adams MA:
Effect of sustained loading on the water content of intervertebral
discs: Implications for disc metabolism. Ann Rheum Dis. 55:880–887.
1996. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Chen J, Baer AE, Paik PY, Yan W and Setton
LA: Matrix protein gene expression in intervertebral disc cells
subjected to altered osmolarity. Biochem Biophys Res Commun.
293:932–938. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Pritchard S, Erickson GR and Guilak F:
Hyperosmotically induced volume change and calcium signaling in
intervertebral disk cells: The role of the actin cytoskeleton.
Biophys J. 83:2502–2510. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Pritchard S and Guilak F: The role of
F-actin in hypo-osmotically induced cell volume change and calcium
signaling in anulus fibrosus cells. Ann Biomed Eng. 32:103–111.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Freemont AJ: The cellular pathobiology of
the degenerate intervertebral disc and discogenic back pain.
Rheumatology (Oxford). 48:5–10. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Johnson NL, Gardner AM, Diener KM,
Lange-Carter CA, Gleavy J, Jarpe MB, Minden A, Karin M, Zon LI and
Johnson GL: Signal transduction pathways regulated by
mitogen-activated/extracellular response kinase kinase kinase
induce cell death. J Biol Chem. 271:3229–3237. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Hiyama A, Gogate SS, Gajghate S, Mochida
J, Shapiro IM and Risbud MV: BMP-2 and TGF-beta stimulate
expression of beta1,3-glucuronosyl transferase 1 (GlcAT-1) in
nucleus pulposus cells through AP1, TonEBP and Sp1: Role of MAPKs.
J Bone Miner Res. 25:1179–1190. 2010.PubMed/NCBI
|
|
26
|
Dong ZH, Wang DC, Liu TT, Li FH, Liu RL,
Wei JW and Zhou CL: The roles of MAPKs in rabbit nucleus pulposus
cell apoptosis induced by high osmolality. Eur Rev Med Pharmacol
Sci. 18:2835–2845. 2014.PubMed/NCBI
|
|
27
|
Li P, Gan Y, Wang H, Xu Y, Li S, Song L,
Zhang C, Ou Y, Wang L and Zhou Q: Role of the ERK1/2 pathway in
osmolarity effects on nucleus pulposus cell apoptosis in a disc
perfusion culture. J Orthop Res. 35:86–92. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Evangelou E, Kerkhof HJ, Styrkarsdottir U,
Ntzani EE, Bos SD, Esko T, Evans DS, Metrustry S, Panoutsopoulou K,
Ramos YF, et al: A meta-analysis of genome-wide association studies
identifies novel variants associated with osteoarthritis of the
hip. Ann Rheum Dis. 73:2130–2136. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Tsezou A: Osteoarthritis year in review
2014: Genetics and genomics. Osteoarthritis Cartilage.
22:2017–2024. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Gee F, Rushton MD, Loughlin J and Reynard
LN: Correlation of the osteoarthritis susceptibility variants that
map to chromosome 20q13 with an expression quantitative trait locus
operating on NCOA3 and with functional variation at the
polymorphism rs116855380. Arthritis Rheumatol. 67:2923–2932. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Pierre S, Bats AS and Coumoul X:
Understanding SOS (Son of Sevenless). Biochem Pharmacol.
82:1049–1056. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Xiao Z, Li L, Li Y, Zhou W, Cheng J, Liu
F, Zheng P, Zhang Y and Che Y: Rasfonin, a novel 2-pyrone
derivative, induces ras-mutated Panc-1 pancreatic tumor cell death
in nude mice. Cell Death Dis. 5:e12412014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Chang YL, Ho BC, Sher S, Yu SL and Yang
PC: miR-146a and miR-370 coordinate enterovirus 71-induced cell
apoptosis through targeting SOS1 and GADD45beta. Cell Microbiol.
17:802–818. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Yue Y, Zhang M, Zhang J, Duan L and Li Z:
SOS1 gene overexpression increased salt tolerance in transgenic
tobacco by maintaining a higher K(+)/Na(+) ratio. J Plant Physiol.
169:255–261. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Ariga H, Katori T, Yoshihara R, Hase Y,
Nozawa S, Narumi I, Iuchi S, Kobayashi M, Tezuka K, Sakata Y, et
al: Arabidopsis sos1 mutant in a salt-tolerant accession revealed
an importance of salt acclimation ability in plant salt tolerance.
Plant Signal Behav. 8:e247792013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Kim JH, Nguyen NH, Jeong CY, Nguyen NT,
Hong SW and Lee H: Loss of the R2R3 MYB, AtMyb73, causes
hyper-induction of the SOS1 and SOS3 genes in response to high
salinity in Arabidopsis. J Plant Physiol. 170:1461–1465. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Katschnig D, Bliek T, Rozema J and Schat
H: Constitutive high-level SOS1 expression and absence of HKT1;1
expression in the salt-accumulating halophyte Salicornia
dolichostachya. Plant Sci. 234:144–154. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Stade K, Ford CS, Guthrie C and Weis K:
Exportin 1 (Crm1p) is an essential nuclear export factor. Cell.
90:1041–1050. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Engel K, Kotlyarov A and Gaestel M:
Leptomycin B-sensitive nuclear export of MAPKAP kinase 2 is
regulated by phosphorylation. EMBO J. 17:3363–3371. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Toone WM, Kuge S, Samuels M, Morgan BA,
Toda T and Jones N: Regulation of the fission yeast transcription
factor Pap1 by oxidative stress: Requirement for the nuclear export
factor Crm1 (Exportin) and the stress-activated MAP kinase
Sty1/Spc1. Genes Dev. 12:1453–1463. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Ferrigno P, Posas F, Koepp D, Saito H and
Silver PA: Regulated nucleo/cytoplasmic exchange of HOG1 MAPK
requires the importin beta homologs NMD5 and XPO1. EMBO J.
17:5606–5614. 1998. View Article : Google Scholar : PubMed/NCBI
|