Introduction
Lung cancer is the leading cause of
cancer-associated mortality worldwide, with ~1.35 million new cases
every year worldwide (1). Between 75
and 80% of lung cancer cases are non-small cell lung cancer
(NSCLC), which has an overall 5-year survival rate of only 10%
(2). Despite novel methods targeting
early diagnosis and recent advancements in treatments, the
prognosis and survival rate of NSCLC patients remains relatively
poor. Between 40 and 50% of patients will eventually succumb to
relapse or metastatic disease after curative resection (3). However, few reliable prognostic
biomarkers are available in clinical practice, particularly in
patients who have undergone curative surgical resection (4). Therefore, there is a requirement to
identify novel prognostic biomarkers that could aid prognosis
prediction and optimize the treatment of NSCLC patients.
MicroRNAs (miRNAs/miRs) are a class of small (19–24
nucleotides in length), highly conserved non-coding RNAs that bind
to the 3′-untranslated regions of target mRNAs and suppress their
translation to proteins (5). It has
become increasingly evident that different miRNAs can play either
oncogenic or tumor-suppressive roles in a wide variety of pathways,
depending on the target genes or the cellular context (6,7). miR-200c
is a member of the miR-200 family, which consists of five members
(miR-200a, miR-200b and miR-429 comprise cluster 1, which is
located on chromosome 1p36; and miR-200c and miR-141 comprise
cluster 2, which is located on chromosome 12p13) (8). Recent investigations have shown that
members of the miR-200 family could promote or repress different
cancer types via various pathways (9,10).
However, studies investigating the association between the
expression level of miR-200c and the clinical outcome in resected
NSCLC patients are few in number and have returned contradictory
results (8,11,12). The
specific role of miR-200c in NSCLC has, therefore, not yet been
elucidated.
In the present study, the expression of miR-200c was
examined in 110 clinical NSCLC samples and the association between
miR-200c expression and variable clinicopathological features and
patient prognosis was analyzed. The present study demonstrated that
high miR-200c expression levels were associated with poor
disease-free and overall survival rates in NSCLC patients after
surgery, and that its presence was an independent prognostic
factor.
Patients and methods
Patients
A total of 110 tumor samples were collected from
patients (65 male, 45 female; mean age, 60.5 years; age range,
41–78, years) who had been pathologically diagnosed with primary
NSCLC and who underwent complete tumor resection (lobectomy or
pneumonectomy) with regional lymph node dissection at the
Department of Thoracic Surgery, Qilu Hospital (Jinan, China)
between January and December 2008.
The study was approved by the Institutional Review
Board at Qilu Hospital and written informed consent was obtained
from all the patients involved in the study. No patients had
undergone preoperative radiotherapy or chemotherapy. The
postsurgical histological type and grade of cancer cell
differentiation was determined by the World Health Organization
classification system (revised in 2004), and the pathological
Tumor-Node-Metastasis (TNM) classification stage was determined by
the 2009 staging system of the Union for International Cancer
Control. The complete follow-up data (until December 2015, loss or
mortality) were included. The clinicopathological characteristics
of these 110 patients are summarized in Table I.
 | Table I.Correlation of clinicopathological
variables with miR-200c expression in NSCLC. |
Table I.
Correlation of clinicopathological
variables with miR-200c expression in NSCLC.
| Clinical
variable | n | Low expression,
% | High expression,
% | P-value |
|---|
| Age, years |
|
|
| 0.051 |
|
<60 | 62 | 30 | 32 |
|
| ≥60 | 48 | 14 | 34 |
|
| Sex |
|
|
| 0.437 |
| Male | 65 | 24 | 41 |
|
|
Female | 45 | 20 | 25 |
|
| Smoking history |
|
|
| 1.000 |
| Yes | 48 | 19 | 29 |
|
| No | 62 | 25 | 37 |
|
| Histology |
|
|
| 0.112 |
|
SCC | 66 | 22 | 44 |
|
|
Adenocarcinoma | 44 | 22 | 22 |
|
|
Differentiation |
|
|
| 0.452 |
|
Well | 28 | 10 | 18 |
|
|
Moderate | 52 | 24 | 28 |
|
|
Poor | 30 | 10 | 20 |
|
| Tumor size, cm |
|
|
| 0.172 |
| ≤3 | 46 | 22 | 24 |
|
|
>3 | 64 | 22 | 42 |
|
| Lymph node
metastasis |
|
|
| 0.000 |
| N0 | 56 | 29 | 27 |
|
|
N1/N2 | 54 | 15 | 39 |
|
| TNM stage |
|
|
| 0.088 |
| I | 46 | 23 | 23 |
|
| II | 45 | 17 | 28 |
|
|
III | 19 | 4 | 15 |
|
Nucleic acid isolation and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
RNA was extracted from fresh-frozen tissues stored
at −80°C. Reverse transcription was performed with reverse
transcription with a Revertaid H Minus First Strand cDNA Synthesis
kit (Tiangen Biotech Co., Ltd., Beijing, China) with miRNA-specific
primers, using 1 µg of total RNA in a 20 µl reverse transcriptase
reaction mixture. miRNA levels were evaluating using SYBR Green PCR
Master Mix (Tiangen) for miR-200c and U6 RNA with stem-loop RT-PCR,
as described previously (13),
following the manufacturer's protocol and using a 20-µl reaction
mixture. The reactions were performed on an ABI 7500 Real-Time PCR
system (Applied Biosystems; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA). Thermocycling conditions were: 95°C for 10 min
followed by 40 cycles of 95°C for 15 sec and 62°C for 1 min.
Expression of the U6 small nuclear RNA was used as an internal
control to normalize all results. The total RNA of the normal
sample was used as a normal control. Data were analyzed using
Sequence Detection Software 1.4 (Applied Biosystems; Thermo Fisher
Scientific, Inc.). The relative quantity of the transcript was
calculated using the 2−∆∆Cq method (14). There were six experimental repeats.
Customized primers were designed using the Primer Express software
(Applied Biosystems; Thermo Fisher Scientific, Inc.). Primer
sequences used are listed in Table
II.
 | Table II.Sequences of the primers of miR-200c
and U6 RNA. |
Table II.
Sequences of the primers of miR-200c
and U6 RNA.
| Primer | miR-200c
(5′-3′) | U6 RNA (5′-3′) |
|---|
| RT |
CTCGTATCCAGTGCAGGGTCCG |
GTGCAGGGTCCGAGGT |
|
|
AGGTATTCGCACTGGATACGAGCCAAAC |
|
| Forward |
GAGCCGTCTTACCCAGCA |
CTCGCTTCGGCAGCACA |
| Reverse |
GTGCAGGGTCCGAGGTAT |
GTGCAGGGTCCGAGGT |
Statistical analysis
All statistical analyses were performed using SPSS
18.0 (SPSS, Inc., Chicago, IL, USA). The associations between
clinical variables and miR-200c expression were analyzed using the
Pearson χ2 test. Survival curves were plotted using the
Kaplan-Meier method and assessed with a log-rank test to identify
significant differences, with mortality due to lung cancer as the
end point. Multivariate Cox regression was used to perform
multivariate survival analysis (5-year disease-free survival and
5-year overall survival). P<0.05 was considered to indicate
statistical significance.
Results
Correlation of miR-200c expression
with clinicopathological factors
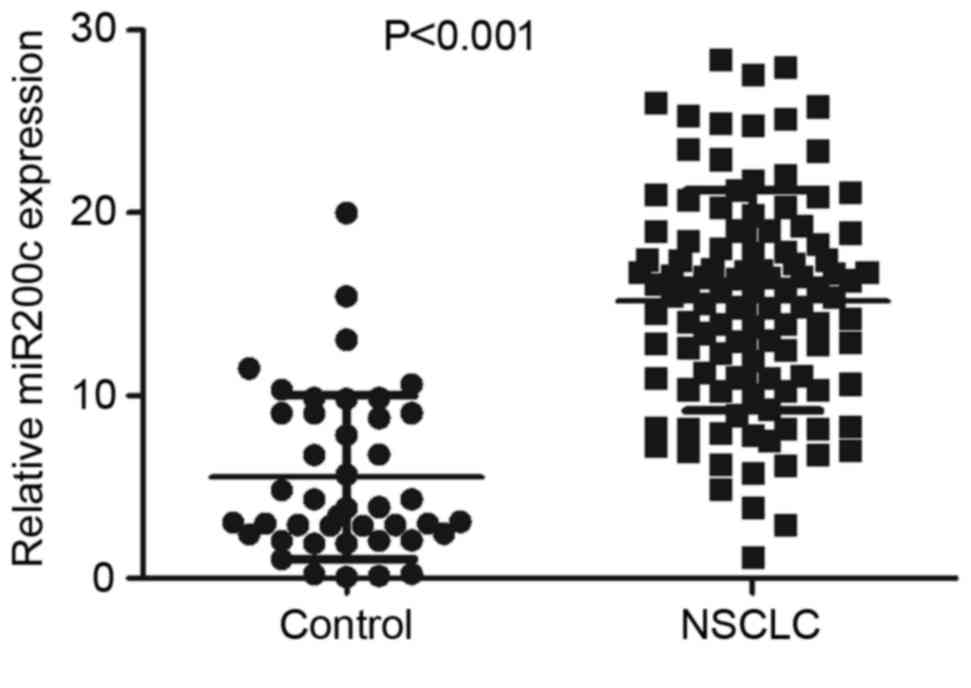
miR-200c levels were quantified by performing
stem-loop RT-PCR on 110 NSCLC specimens and 43 normal lung tissues.
qPCR confirmed that, compared with their corresponding normal
tissues, the NSCLC specimens exhibited upregulated miR-200c
expression (Table III; Fig. 1).
 | Table III.Expression of miR200c in the 110
NSCLC patients and 43 healthy controls. |
Table III.
Expression of miR200c in the 110
NSCLC patients and 43 healthy controls.
| Group | Subjects, n | miR200c expression,
%a | P-value |
|---|
| NSCLC patients | 110 | 15.203±0.575 | <0.001 |
| Healthy
control | 43 | 5.533±0.684 |
|
The association of miR-200c expression with
clinicopathological factors was examined using a χ2
test. Higher miR-200c expression was significantly associated with
positive lymph node metastasis (P<0.001); there was no
statistical significance in the associations between miR-200c
expression and other clinicopathological variables (P>0.05)
(Table I).
Univariate survival analysis for
5-year disease-free survival and 5-year overall survival
Of the 110 NSCLC patients examined in this study,
tumor relapse developed in 89 (80.9%) within the follow-up period:
Local recurrence occurred in 22 patients, distant metastasis in 46
patients, and local recurrence and distant metastasis in 21
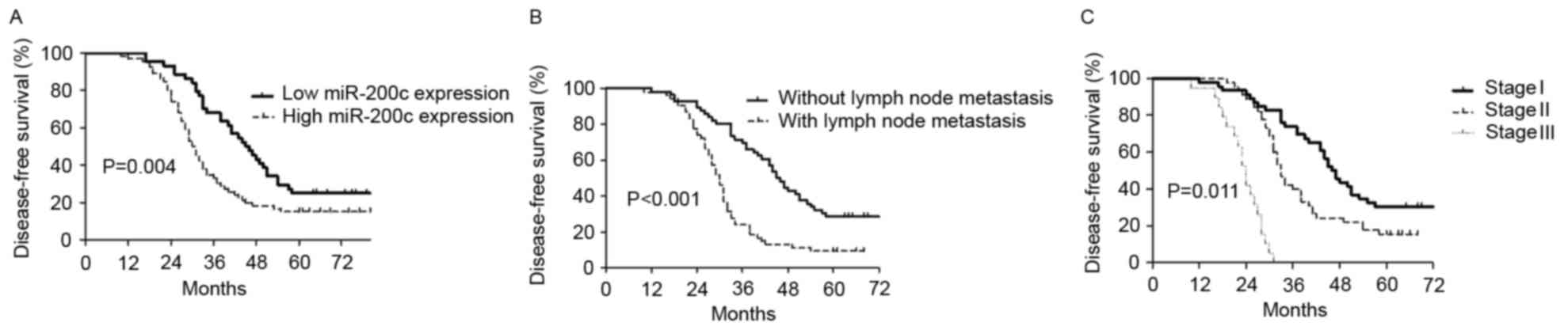
patients. Univariate analysis (log-rank test) demonstrated that
higher miR-200c expression (15.2 vs. 25.0%, P=0.004; Fig. 2A), positive lymph node metastasis (9.3
vs. 28.6%, P<0.001; Fig. 2B) and
advanced TNM stage (0 vs. 15.6 vs. 30.4% for stage III, II, and I,
respectively; P=0.011; Fig. 2C)
significantly predicted decreased 5-year disease-free survival
rates.
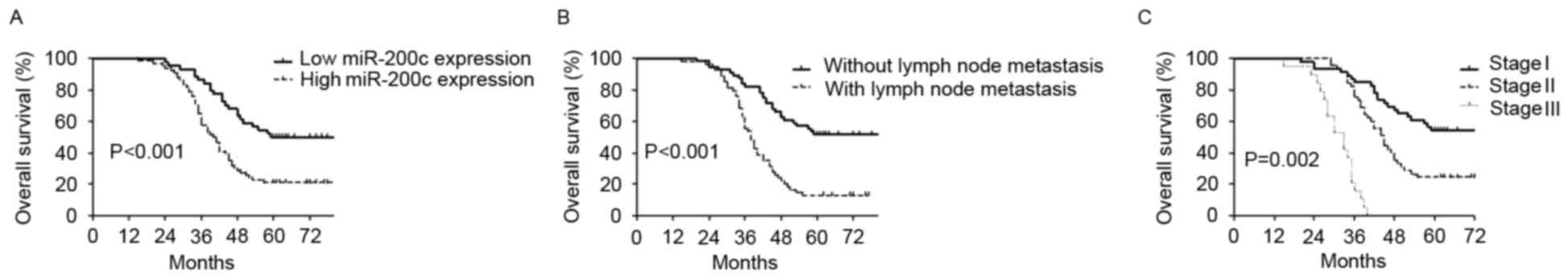
Of the 110 NSCLC patients, 74 (67.3%) succumbed to
cancer-associated causes within 5 years of surgery, and the 5-year
overall survival was 32.7%. Univariate analysis (log-rank test)
demonstrated that high miR-200c expression (21.2 vs. 50.0%,
P<0.001; Fig. 3A), positive lymph
node metastasis (13.0 vs. 51.8%, P<0.001; Fig. 3B) and advanced TNM stage (0 vs. 24.4
vs. 54.3% for stage III, II and I, respectively; P=0.002; Fig. 3C) significantly predicted poor 5-year
overall survival rates (Table
IV).
 | Table IV.Univariate survival analysis for
disease-free survival and overall survival rates. |
Table IV.
Univariate survival analysis for
disease-free survival and overall survival rates.
| Variable | Disease-free
survival rate, P-value | Overall survival
rate, P-value |
|---|
| Age (≤60 vs. >60
years) | 0.406 | 0.739 |
| Sex (male vs.
female) | 0.146 | 0.079 |
| Smoking (yes vs.
no) | 0.769 | 0.330 |
| Histology | 0.429 | 0.844 |
| (SCC vs.
adenocarcinoma) |
|
|
|
Differentiation | 0.911 | 0.094 |
| (poor vs. moderate
vs. well) |
|
|
| Tumor size (≤3 vs.
>3 cm) | 0.314 | 0.953 |
| Lymph node
metastasis | 0.000 | 0.000 |
| (N0 vs. N1/N2) |
|
|
| TNM (stage I vs.
stage II/III) | 0.011 | 0.002 |
| miR200c (high vs.
low) | 0.004 | 0.000 |
Multivariate survival analysis for
5-year disease-free survival and 5-year overall survival
Among all variables, there existed statistical
significance for the association between lymph node metastasis, TNM
stage and miR-200c expression in univariate survival analysis.
Thus, these three variables were assessed using multivariate
survival analysis. The results of multivariate Cox regression
analysis showed that TNM stage (both P<0.000; Table V) and miR-200c expression (P=0.030 and
P=0.006; Table V) retained
significance as independent prognostic factors for unfavorable
5-year disease-free survival and poor 5-year overall survival
rates, respectively.
 | Table V.Multivariate survival analysis for
disease-free survival and overall survival. |
Table V.
Multivariate survival analysis for
disease-free survival and overall survival.
|
| Disease-free
survival | Overall
survival |
|---|
|
|
|
|
|---|
| Variable | 95% CI | Exp(B) | P-value | 95% CI | Exp(B) | P-value |
|---|
| Lymph node
metastasis (N0 vs. N1/N2) | 0.510–1.626 | 0.910 | 0.751 | 0.453–1.661 | 0.868 | 0.668 |
| TNM (stage I vs.
stage II and III) | 1.857–4.661 | 2.942 | 0.000 | 2.177–5.886 | 3.580 | 0.000 |
| miR-200c (high vs.
low) | 1.049–2.585 | 1.647 | 0.030 | 1.241–3.536 | 2.095 | 0.006 |
Discussion
In the clinic, the prognosis of patients with NSCLC
is markedly different, even if they have the same pathological
staging (15,16). This difference may be due to the fact
that patients are in a different disease stage when they are
diagnosed, meaning the current staging system is not sufficient to
predict prognosis and/or inform a treatment strategy for numerous
patients. Therefore, it is necessary to identify and employ novel
biomarkers as possible therapeutic targets or prognostic
predictors, which will be used as an adjunct to the staging system
and contribute to the optimization of treatment for patients with
NSCLC. miRNAs as a recent focus in tumor research serve an
oncogenic or tumor-suppressive role in different cancer types via
various pathways (17–19). miR-200c is a member of the miR-200
family, which is located on chromosome 12p13 and is closely
associated with carcinogenesis and disease progression in a wide
range of cancer types (20,21). To the best of our knowledge, there
have been only 4 clinical studies on miR-200c expression in NSCLC
to date, producing contradictory results (i.e. that miR-200c has
been reported to have oncogenic or tumor-suppressive functions)
(10,12,22,23). The
present study aimed to assess miR-200c expression in NSCLC, and to
investigate the role of miR-200c in relation to carcinogenesis and
the prognosis of NSCLC patients.
The present study demonstrated that miR200c
overexpression was common in NSCLC tissues and significantly
associated with lymph node metastasis. For 5-year disease-free
survival and overall survival rates, Kaplan-Meier analysis showed
that patients with lymph node metastasis, advanced TNM stage and
high miR-200c expression had a poor prognosis. To ascertain whether
the impact of mixed factors was associated with prognosis, Cox
regression multivariate analysis was performed and demonstrated
that only TNM stage and high miR-200c expression had value as
independent prognostic factors. The present results indicate that
TNM stage (which is already internationally recognized) and miR200c
are useful diagnostic markers that may themselves promote tumor
progression in NSCLC and other cancer types.
However, tumor carcinogenesis is a complex process,
in which regulation of cell growth and differentiation must be
altered (24). Genetic and epigenetic
changes can occur at multiple levels, from chromothripsis or the
loss or gain of entire chromosomes to a point mutation that alters
a single DNA nucleotide, or to the silencing or activation of an
miRNA that can alter the expression of up to 500 genes (25,26). To
date, the mechanism through which miR-200c can affect the
carcinogenic potential of cancer cells remains unknown and requires
further elucidation at the molecular level.
The present study indicated that miR-200c is
associated with lymph node metastasis, suggesting that miR-200c may
be upregulated in the metastatic process. However, miRNA-200c has
been shown to restrict the epithelial-mesenchymal transition (EMT)
and metastasis through direct targeting of the cell adhesion
pathway (27,28), particularly the zinc finger E-box
binding homeobox (ZEB)-cadherin 1 axis (29). However, the pleiotropic effect of
miR-200c in the metastatic process is contradictory in in
vitro and in vivo studies (30). A previous study demonstrated that the
metastatic potential of tumor cells could be increased by the
overexpression of miR-200c (31). The
overexpression of miR-200c in a xenograft model was found to
associate with a higher metastatic potential and increased
metastatic colonization (32). The
present findings, in which expression of miR-200c was upregulated
in tumors and associated with poor prognosis and lymph node
metastasis, are consistent with experimental data in ovarian
(33), colorectal (34), gastric (35,36) and
breast (37) cancer. The mechanism
may involve miR-200c overexpression and an increase in metastatic
risk by repressing the expression of E-cadherin transcriptional
repressors ZEB1 and ZEB2, and the final induction of EMT.
In summary, the present study provides evidence that
miR-200c expression in lung cancer tissue may be an effective
predictor for monitoring cancer progression and informing on future
prognosis, although the underlying mechanism remains unclear.
Individual miRNAs can direct different biological processes by
regulating the expression of multiple downstream targets, including
oncogenes and tumor suppressor genes, which suggests that the
contributions of miRNAs to tumorigenesis vary between different
tumors. The present results could provide important information for
predicting prognosis and tumor progression. However, more
well-designed studies with larger sample sizes and a standardized
methodology, investigating associated candidate target genes, are
required.
References
|
1
|
Sibille A, Paulus A, Martin M, Bourhaba M,
Barthélemy N, Radermecker M, Corhay JL, Louis R and Duysinx B:
Management of non-small cell lung cancer. Rev Med Liege.
70:432–441. 2015.(In French). PubMed/NCBI
|
|
2
|
Venkatesulu BP, Mallick S, Singh A and
Julka PK: Non small cell carcinoma of lung with metachronous breast
metastasis and cardiac tamponade: Unusual presentation of a common
cancer. J Egypt Natl Canc Inst. 27:165–169. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Sun Z, Wang Z, Liu X and Wang D: New
development of inhibitors targeting the PI3K/AKT/mTOR pathway in
personalized treatment of non-small-cell lung cancer. Anticancer
Drugs. 26:1–14. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wu CF, Fu JY, Yeh CJ, Liu YH, Hsieh MJ, Wu
YC, Wu CY, Tsai YH and Chou WC: Recurrence risk factors analysis
for stage I non-small cell lung cancer. Medicine (Baltimore).
94:e13372015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Feng B, Zhang K, Wang R and Chen L:
Non-small-cell lung cancer and miRNAs: Novel biomarkers and
promising tools for treatment. Clin Sci (Lond). 128:619–634. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ghasemkhani N, Shadvar S, Masoudi Y,
Talaei AJ, Yahaghi E, Goudarzi PK and Shakiba E: Down-regulated
MicroRNA 148b expression as predictive biomarker and its prognostic
significance associated with clinicopathological features in
non-small-cell lung cancer patients. Diagn Pathol. 10:1642015.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Bai T, Dong DS and Pei L: Synergistic
antitumor activity of resveratrol and miR-200c in human lung
cancer. Oncol Rep. 31:2293–2297. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Shi L, Zhang S, Wu H, Zhang L, Dai X, Hu
J, Xue J, Liu T, Liang Y and Wu G: MiR-200c increases the
radiosensitivity of non-small-cell lung cancer cell line A549 by
targeting VEGF-VEGFR2 pathway. PLoS One. 8:e783442013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Li J, Li X, Ren S, Chen X, Zhang Y, Zhou
F, Zhao M, Zhao C, Chen X, Cheng N, et al: miR-200c overexpression
is associated with better efficacy of EGFR-TKIs in non-small cell
lung cancer patients with EGFR wild-type. Oncotarget. 5:7902–7916.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Liu XG, Zhu WY, Huang YY, Ma LN, Zhou SQ,
Wang YK, Zeng F, Zhou JH and Zhang YK: High expression of serum
miR-21 and tumor miR-200c associated with poor prognosis in
patients with lung cancer. Med Oncol. 29:618–626. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kim MK, Jung SB, Kim JS, Roh MS, Lee JH,
Lee EH and Lee HW: Expression of microRNA miR-126 and miR-200c is
associated with prognosis in patients with non-small cell lung
cancer. Virchows Arch. 465:463–471. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Li X, Li D, Zhuang Y, Shi Q, Wei W and Ju
X: Overexpression of miR-708 and its targets in the childhood
common precursor B-cell ALL. Pediatr Blood Cancer. 60:2060–2067.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Hansen O, Schytte T, Nielsen M and Brink
C: Age dependent prognosis in concurrent chemo-radiation of locally
advanced NSCLC. Acta Oncol. 54:333–339. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Strøm HH, Bremnes RM, Sundstrøm SH,
Helbekkmo N and Aasebø U: Poor prognosis patients with inoperable
locally advanced NSCLC and large tumors benefit from palliative
chemoradiotherapy: A subset analysis from a randomized clinical
phase III trial. J Thorac Oncol. 9:825–833. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Liu CM, Peng CY, Liao YW, Lu MY, Tsai ML,
Yeh JC, Yu CH and Yu CC: Sulforaphane targets cancer stemness and
tumor initiating properties in oral squamous cell carcinomas via
miR-200c induction. J Formos Med Assoc. 116:41–48. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Xie Y, Wehrkamp CJ, Li J, Wang Y, Wang Y,
Mott JL and Oupický D: Delivery of miR-200c Mimic with Poly (amido
amine) CXCR4 antagonists for combined inhibition of
cholangiocarcinoma cell invasiveness. Mol Pharm. 13:1073–1080.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhang H, Liu J, Qu D, Wang L, Luo JY, Lau
CW, Liu P, Gao Z, Tipoe GL, Lee HK, et al: Inhibition of miR-200c
restores endothelial function in diabetic mice through suppression
of COX-2. Diabetes. 65:1196–1207. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Mutlu M, Raza U, Saatci Ö, Eyüpoğlu E,
Yurdusev E and Şahin Ö: MiR-200c: A versatile watchdog in cancer
progression, EMT, and drug resistance. J Mol Med (Berl).
94:629–644. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Dimri M, Kang M and Dimri GP: A
miR-200c/141-BMI1 autoregulatory loop regulates oncogenic activity
of BMI1 in cancer cells. Oncotarget. 7:36220–36234. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ceppi P, Mudduluru G, Kumarswamy R, Rapa
I, Scagliotti GV, Papotti M and Allgayer H: Loss of miR-200c
expression induces an aggressive, invasive, and chemoresistant
phenotype in non-small cell lung cancer. Mol Cancer Res.
8:1207–1216. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Tejero R, Navarro A, Campayo M, Viñolas N,
Marrades RM, Cordeiro A, Ruíz-Martínez M, Santasusagna S, Molins L,
Ramirez J and Monzó M: miR-141 and miR-200c as markers of overall
survival in early stage non-small cell lung cancer adenocarcinoma.
PLoS One. 9:e1018992014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Brücher BL and Jamall IS: Cell-cell
communication in the tumor microenvironment, carcinogenesis, and
anticancer treatment. Cell Physiol Biochem. 34:213–243. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wang L, Li G, Yao ZQ, Moorman JP and Ning
S: MicroRNA regulation of viral immunity, latency, and
carcinogenesis of selected tumor viruses and HIV. Rev Med Virol.
25:320–341. 2015. View
Article : Google Scholar : PubMed/NCBI
|
|
26
|
Bizzarri M and Cucina A: Tumor and the
microenvironment: A chance to reframe the paradigm of
carcinogenesis? Biomed Res Int. 2014:9340382014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Liu S, Tetzlaff MT, Wang T, Yang R, Xie L,
Zhang G, Krepler C, Xiao M, Beqiri M, Xu W, et al: miR-200c/Bmi1
axis and epithelial-mesenchymal transition contribute to acquired
resistance to BRAF inhibitor treatment. Pigment Cell Melanoma Res.
28:431–441. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Tamagawa S, Beder LB, Hotomi M, Gunduz M,
Yata K, Grenman R and Yamanaka N: Role of miR-200c/miR-141 in the
regulation of epithelial-mesenchymal transition and migration in
head and neck squamous cell carcinoma. Int J Mol Med. 33:879–886.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Rajabi H, Alam M, Takahashi H, Kharbanda
A, Guha M, Ahmad R and Kufe D: MUC1-C oncoprotein activates the
ZEB1/miR-200c regulatory loop and epithelial-mesenchymal
transition. Oncogene. 33:1680–1689. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Knezevic J, Pfefferle AD, Petrovic I,
Greene SB, Perou CM and Rosen JM: Expression of miR-200c in
claudin-low breast cancer alters stem cell functionality, enhances
chemosensitivity and reduces metastatic potential. Oncogene.
34:5997–6006. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Gravgaard KH, Lyng MB, Laenkholm AV,
Søkilde R, Nielsen BS, Litman T and Ditzel HJ: The miRNA-200 family
and miRNA-9 exhibit differential expression in primary versus
corresponding metastatic tissue in breast cancer. Breast Cancer Res
Treat. 134:207–217. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Korpal M, Ell BJ, Buffa FM, Ibrahim T,
Blanco MA, Celià-Terrassa T, Mercatali L, Khan Z, Goodarzi H, Hua
Y, et al: Direct targeting of Sec23a by miR-200s influences cancer
cell secretome and promotes metastatic colonization. Nat Med.
17:1101–1108. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
33
|
Vilming Elgaaen B, Olstad OK, Haug KB,
Brusletto B, Sandvik L, Staff AC, Gautvik KM and Davidson B: Global
miRNA expression analysis of serous and clear cell ovarian
carcinomas identifies differentially expressed miRNAs including
miR-200c-3p as a prognostic marker. BMC Cancer. 14:802014.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Toiyama Y, Hur K, Tanaka K, Inoue Y,
Kusunoki M, Boland CR and Goel A: Serum miR-200c is a novel
prognostic and metastasis-predictive biomarker in patients with
colorectal cancer. Ann Surg. 259:735–743. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zhang HP, Sun FB and Li SJ: Serum miR-200c
expression level as a prognostic biomarker for gastric cancer.
Genet Mol Res. 14:15913–15920. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Valladares-Ayerbes M, Reboredo M,
Medina-Villaamil V, Iglesias-Díaz P, Lorenzo-Patiño MJ, Haz M,
Santamarina I, Blanco M, Fernández-Tajes J, Quindós M, et al:
Circulating miR-200c as a diagnostic and prognostic biomarker for
gastric cancer. J Transl Med. 10:1862012. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Antolin S, Calvo L, Blanco-Calvo M,
Santiago MP, Lorenzo-Patiño MJ, Haz-Conde M, Santamarina I,
Figueroa A, Antón-Aparicio LM and Valladares-Ayerbes M: Circulating
miR-200c and miR-141 and outcomes in patients with breast cancer.
BMC Cancer. 15:2972015. View Article : Google Scholar : PubMed/NCBI
|