Introduction
Among all major cancers, prostate cancer (PCa) has
one of the worst prognoses; it was ranked 2nd for cancer-associated
mortality causes worldwide in 2012 (1). There is evidence to suggest that this
may be due to carcinoma in the prostate being more inclined to
metastasize (2). The lymph nodes and
bone are the destination of metastatic cells in 70–80% cases of
prostate cancer-associated mortality (3). However, early stage diagnosis methods
currently available for PCa are poor, and the lack of effective
therapies for advanced carcinoma results in a high mortality rate
among patients.
Notch1 is a type I transmembrane protein, which has
a dual function of cell membrane surface receptors and nuclear
transcription regulation (4).
Previous studies have reported that the Notch signaling pathway may
be aberrantly activated, contributing to the development, invasion
and metastasis of a wide variety of human cancers, including
cervical, lung, colon, head and neck, renal carcinoma, acute
myeloid, Hodgkin and large-cell lymphomas and pancreatic cancer
(5–7).
Notch are a family of transmembrane proteins with epidermal growth
factor-like domains. There are four Notch receptors (Notch-1 to
−4), as well as six ligands (Jagged1 and 2 and d-like 1, 2, 3 and
4), in mammalian cells (8,9). The signaling pathway is divided into an
extracellular and intracellular region, the latter of which may be
cleaved when the associated genes are expressed (10–12).
It is established that multiple steps are involved
in the progression of metastasis in prostatic tumors, including
angiogenesis, migration and intravasation, leading to tumor cell
invasion and ultimately, metastasis (13). It has been reported that Notch
signaling may regulate the metastasis of tumors in multiple tissues
and organs (14–17). Therefore, in the present study, Notch1
was knocked down to investigate the function and mechanism of
Notch1 in the invasion and metastasis of human PCa LNCaP cells
in vitro.
Materials and methods
Cell culture
The human prostatic carcinoma LNCaP, PC-3 and DU 145
cell lines and the immortalized human prostatic epithelial RWPE-1
cell line were obtained from the American Type Culture Collection
(Manassas, VA, USA). LNCaP, PC-3 and DU 145 cells were maintained
in RPMI-1640 medium (Cellgro; Corning Life Sciences, Corning, NY,
USA) supplemented with 10% fetal bovine serum (FBS; Biowest USA,
Riverside, MO, USA). RWPE-1 cells were maintained in keratinocyte
serum-free medium (Invitrogen; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA). All cells were incubated in an environment of 5%
CO2 at 37°C.
Western blot analysis
LNCaP, PC-3, DU 145 and RWPE-1 cells were collected
and washed twice with cold PBS, and then lysed in
radioimmunoprecipitation assay buffer (Beyotime Institute of
Biotechnology, Jiangsu, China) containing the protease inhibitor
phenylmethanesulfonyl fluoride, with mild sonication on ice. Cell
lysates (20 µg) were used for western blot analysis, subsequent to
11.5% SDS-PAGE, with primary antibodies against Notch1 (cat. no.
3608S), cleaved-Notch1 (cat. no. 4147S) and GAPDH (cat. no. 5174),
and a secondary HRP-conjugated anti-rabbit IgG (cat. no. 7074S).
All antibodies were used at a 1:2,000 dilution and were purchased
from Cell Signaling Technology, Inc. (Danvers, MA, USA).
Non-specific binding was blocked by incubating with tris-buffered
saline with Tween-20 and 5% skimmed milk powder for 60 min at room
temperature. The membrane was incubated with the primary antibodies
at 4°C overnight, then with the secondary antibody for 2 h at room
temperature. The band intensities of the target proteins and GAPDH
were quantified using Image Lab 5.0 software (Bio-Rad Laboratories,
Inc., Hercules, CA, USA), and the intensity of each target protein
was normalized to the corresponding GAPDH level.
RNA interference
In a 6-well tissue culture plate, LNCaP cells at
50–70% confluency in 800 µl of short hairpin RNA (shRNA) plasmid
transfection medium (cat. no. sc-108062) was transfected with 200
µl Notch1 shRNA plasmid (cat. no. sc-36095-SH) or control shRNA
plasmid-A/shRNA plasmid transfection reagent complex (cat. no.
sc-108060), all from Santa Cruz Biotechnology, Inc. (Dallas, TX,
USA). Following transfection for 24 h at 37°C, RPMI medium was
prepared containing twice the usual concentration of FBS (20%) and
antibiotics. A total of 1 ml prepared medium was added to each
well, and the cells were incubated for an another 24 h at 37°C with
5% CO2. The medium was then replaced with RPMI with 10%
FBS containing 10 µg/ml of puromycin. Every 2–3 days, medium was
aspirated out and replaced with freshly prepared RPMI with 10% FBS.
The cells were subsequently collected and processed for western
blotting, reverse transcription-quantitative polymerase chain
reaction (RT-qPCR), cell invasion and proliferation assays.
RT-qPCR
RNA was extracted from the cell lines using TRIzol
(Thermo Fisher Scientific, Inc.). RNA (1–2 µg) was reverse
transcribed to cDNA using the Transcription First Strand cDNA
Synthesis kit (Roche Diagnostics, Basel, Switzerland). qPCR
analyses were performed using Fast Start Universal SYBR-Green
Master (ROX; Roche Diagnostics) on the ABI 7500FAST system (Applied
Biosystems; Thermo Fisher Scientific, Inc.) The thermocycling
settings were as follows: An initial temperature of 95°C for 5 min;
then 35 cycles of 95°C for 15 sec, 60°C for 60 sec, and 72°C for 60
sec; then a final extension step of 10 min at 72°C. qPCR results
for target gene expression were normalized using GAPDH as the
internal control. Relative target gene expression levels normalized
to GAPDH were determined with the formula 2−∆Cq, in
which ∆Cq=Cqtargetgene-CqGAPDH. To calculate
the fold changes of target gene expression in LNCaP cells
transfected with Notch1 shRNA plasmid compared with the control
cells, the 2−∆∆Cq method was used (17), in which ∆∆Cq=∆CqNotch1
shRNA-∆Cqcell control, and log2 values
were calculated. The sequences for primers used in the RT-qPCR
assay, as supplied by Invitrogen (Thermo Fisher Scientific, Inc.)
are listed in Table I.
 | Table I.Sequences for primers used for reverse
transcription-quantitative polymerase chain reaction analysis. |
Table I.
Sequences for primers used for reverse
transcription-quantitative polymerase chain reaction analysis.
| Gene symbol | Sequence |
|---|
| Notch1 | Forward
5′-GAGGCGTGGCAGACTATGC-3′ |
|
| Reverse
5′-CTTGTACTCCGTCAGCGTGA-3′ |
| MTA1 | Forward
5′-ACGCAACCCTGTCAGTCTG-3′ |
|
| Reverse
5′-GGGCAGGTCCACCATTTCC-3′ |
| KISS-1 | Forward
5′-AGCAGCTAGAATCCCTGGG-3′ |
|
| Reverse
5′-AGGCCGAAGGAGTTCCAGT-3′ |
| MKK4 | Forward
5′-TGAGAAGGGTGACTGCATCG-3′ |
|
| Reverse
5′-ACCAAACCATTGACACCGAAG-3′ |
| KAI1 | Forward
5′-TGTCCTGCAAACCTCCTCCA-3′ |
|
| Reverse
5′-CCATGAGCATAGTGACTGCCC-3′ |
| GAPDH | Forward
5′-GGCTGAGAACGGGAAGCTTGTCAT-3′ |
|
| Reverse
5′-CAGCCTTCTCCATGGTGGTGAAGA-3′ |
Invasion assay
Matrigel (BD Biosciences, Franklin Lakes, NJ, USA)
was thawed at 4°C overnight and then diluted 1:4 in serum free-cold
cell culture media. Matrigel (50 µl) was added to the upper
chambers of a 24-well Transwell plate, and the insert was gently
rotated to ensure the entire membrane was coated. Subsequently, the
Transwell was incubated at 37°C for 5 h for gelling and to absorb
the residual liquid. LNCaP cells transfected with Notch1 shRNA
plasmid or control shRNA plasmid-A were then starved for 24 h in
serum-free RPMI containing 1% FBS, and cells from the tissue
culture flasks were harvested using trypsin/EDTA. The cells were
then washed once with serum-free RPMI, and resuspended in
serum-free RPMI at a density of 1×106 cells/ml. A total
of 100 µl cell suspension was added to the Matrigel, the lower
chamber of the Transwell plate was filled with 600 µl cell culture
media containing 10% FBS, and the plate was incubated at 37°C for
24 h. Crystal violet dye (as 0.09% crystal violet in 10% ethanol)
was then prepared and 500 µl dye was added to the empty wells. The
chamber was moved with forceps into the dye well, incubated in the
dye for 30 min at room temperature, washed in water for ~5 sec, and
the inner surface was then cleaned with a cotton swab to remove the
dye that remained inside. Visual/qualitative observations were then
made using a microscope at ×100 magnification, and the numbers of
invading cells were manually counted. All assays were performed in
triplicate for 3 independent experiments.
Cell proliferation assay
Cell proliferation analysis was performed using the
WST-1 assay, a colorimetric assay for the nonradioactive
quantification of cell proliferation, cell viability and
cytotoxicity (Roche Diagnostics), according to the manufacturer's
protocol. Briefly, cells were plated on 96-well plates (5,000
cells/well) and cell viability was then determined.
Statistical analysis
Data were expressed as the mean ± standard
deviation. Statistical significance of the differences between
groups was analyzed by one-way analysis of variance followed by
Newman-Keuls multiple comparisons tests, using SPSS 10.0 for
windows (SPSS, Inc., Chicago, IL, USA). P<0.05 was considered to
indicate a statistically significant difference.
Results
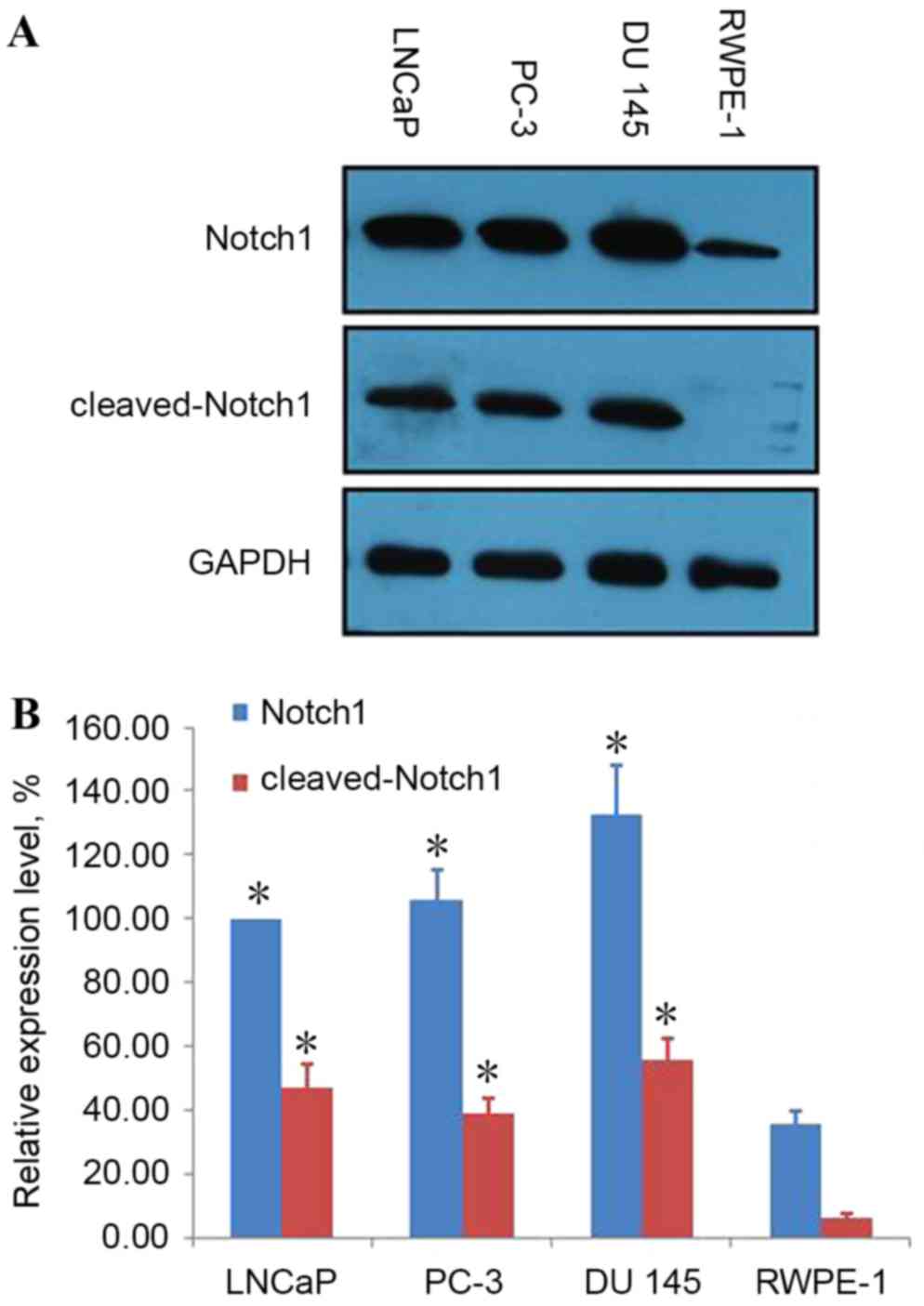
Notch1 and cleaved-Notch1 are
overexpressed in human PCa cells
It was previously reported that Notch signaling may
markedly impact prostate development and disease (18). In the present study, western blot
analysis was employed to detect Notch1 and cleaved-Notch1
expression in the human prostatic carcinoma cell lines LNCaP, PC-3
and DU 145 and the immortalized human RWPE-1 prostatic epithelial
cell line. Notch1 was revealed to be highly expressed in LNCaP,
PC-3 and DU 145 cells compared with RWPE-1 cells, while
cleaved-Notch1 was expressed in LNCaP, PC-3 and DU 145 cells, and
only to a minimal extent in RWPE-1 cells (Fig. 1).
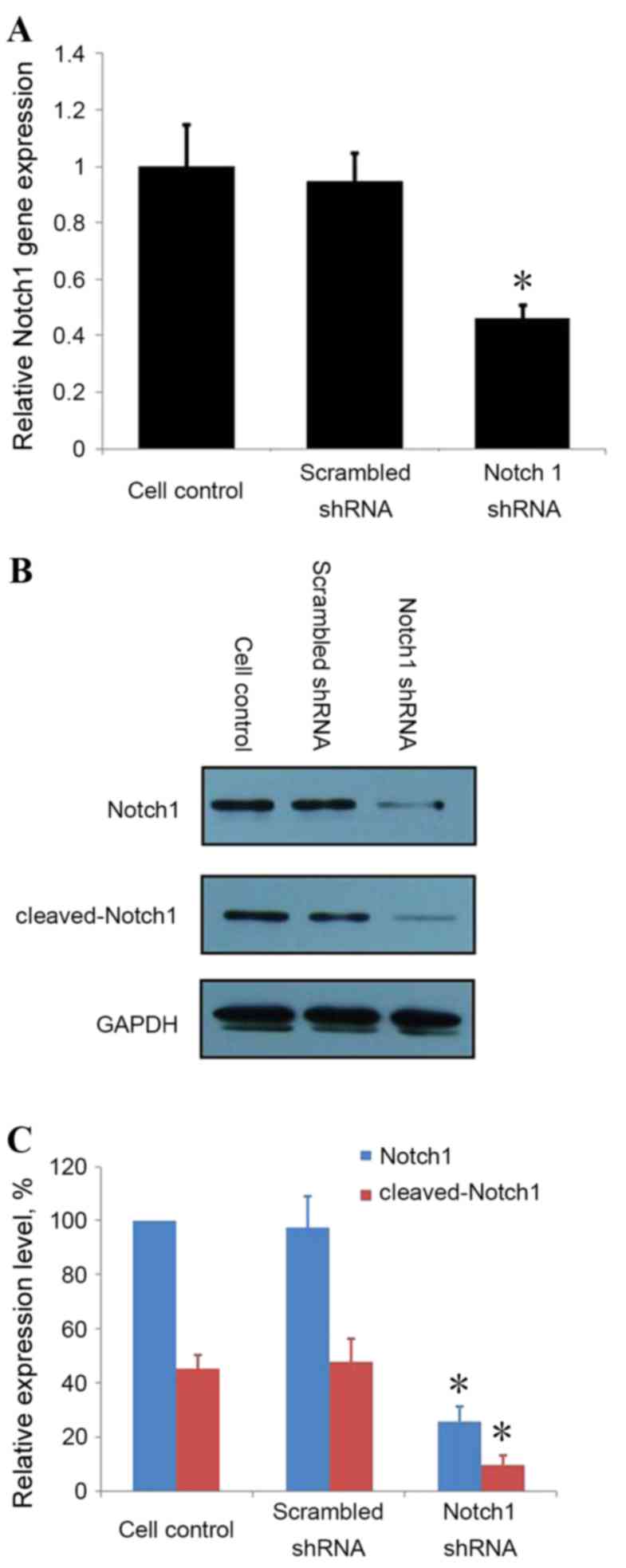
Notch1-knockdown by shRNA in LNCaP
cells
To address the involvement of Notch1 in PCa
invasion, knockdown of Notch1 was achieved by transfecting LNCaP
cells with Notch1 shRNA and scrambled control shRNA. RT-qPCR
revealed Notch1-knockdown in cells transfected with Notch1 shRNA,
but not in the non-transfected control cells or cells transfected
with scrambled shRNA (Fig. 2). These
results were verified by western blot analysis; Notch1 protein and
cleaved-Notch1 expression were downregulated in cells transfected
with Notch1 shRNA compared with non-transfected control cells or
cells transfected with scrambled shRNA (Fig. 2).
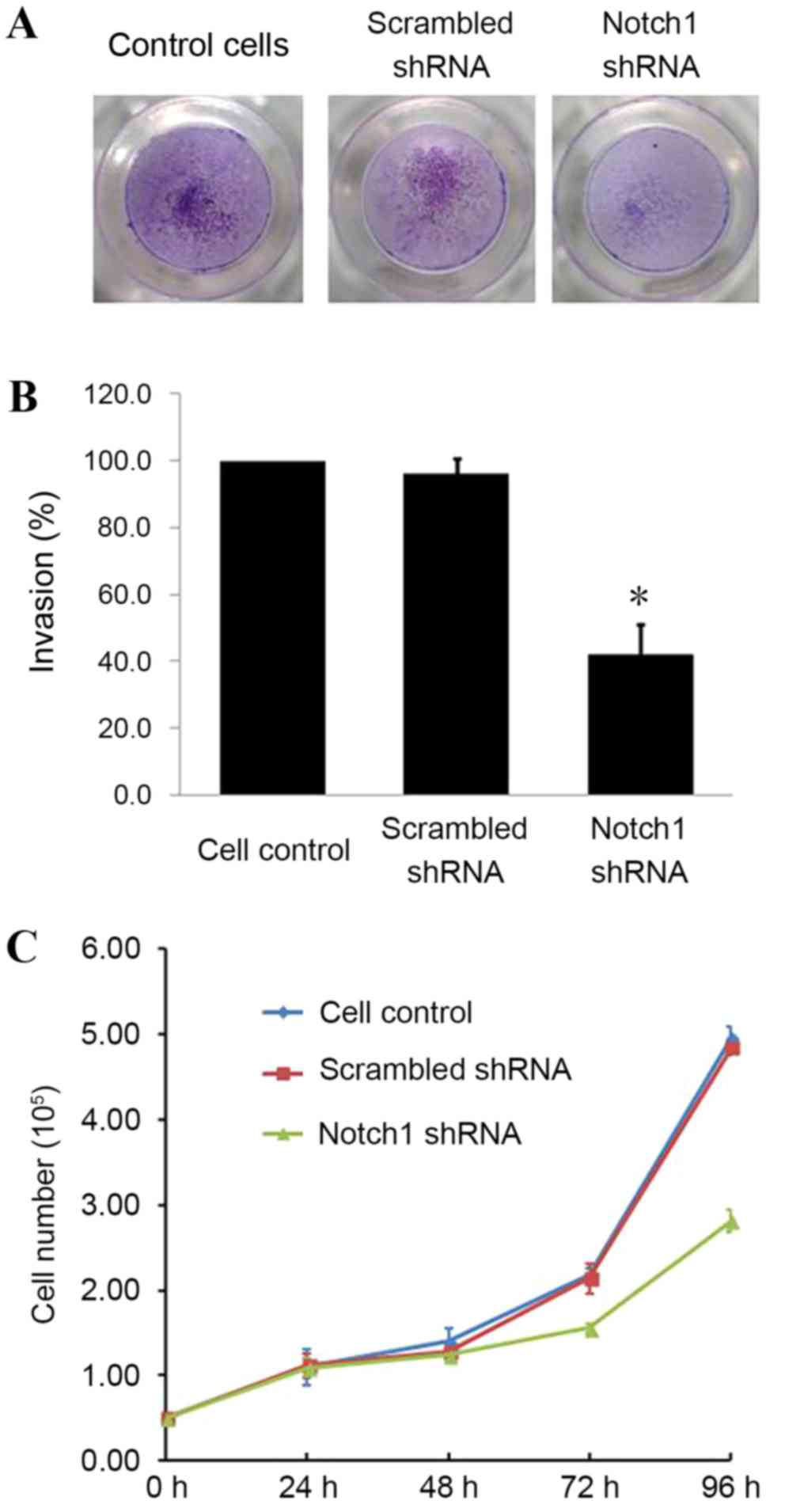
Notch1-knockdown decreases invasion
and proliferation of LNCaP cells
To examine the effects of Notch1-knockdown, LNCaP
cells were subjected to cell invasion and proliferation assays. The
invasion of Notch1-knockdown cells through the extracellular matrix
was reduced compared with non-transfected cells or cells
transfected with scrambled shRNA (Fig.
3A). Notch1-knockdown caused a 60% decrease in cell invasion
(Fig. 3B), indicating that Notch1
conferred invasive properties to PCa cells. In addition, the
proliferation of Notch1-knockdown cells was significantly reduced
48 h post-transfection (Fig. 3C).
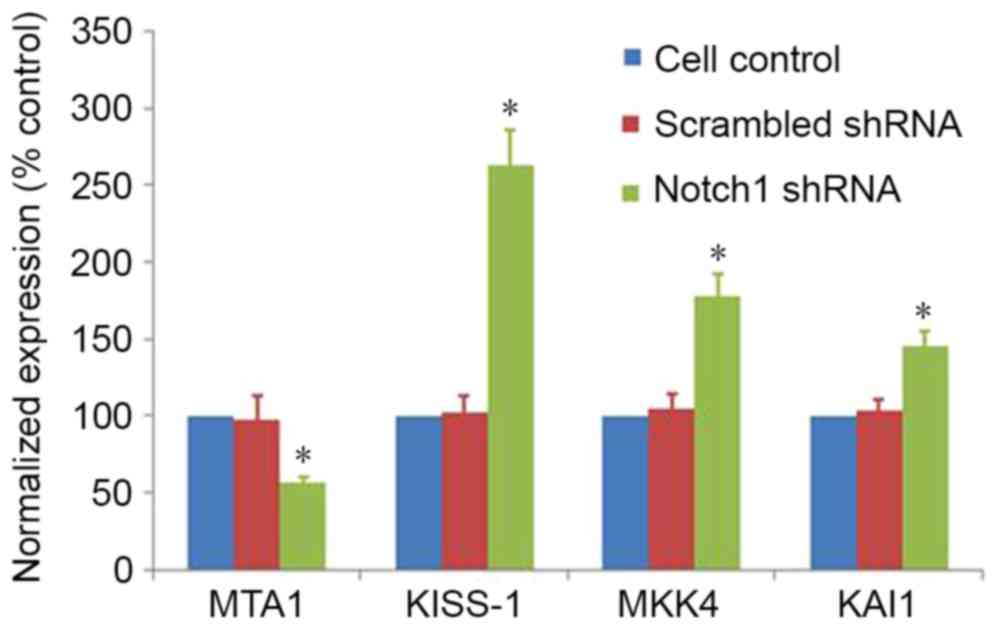
Notch1-knockdown changes the
expression of genes involved in cell invasion
To define the mechanism of Notch1 in LNCaP cells
invasion, RT-qPCR was performed on a number of invasion-associated
genes. Notch1-knockdown resulted in a significant decrease in the
expression of metastasis-associated 1 (MTA1) and increase of KiSS-1
metastasis-suppressor (KISS-1), mitogen-activated protein kinase 4
(MKK4) and cluster of differentiation 82 (KAI1) compared with
non-transfected control cells or cells transfected with scrambled
shRNA (Fig. 4). These results
indicated that Notch1 may regulate the expression of these
downstream target genes that are involved in extracellular matrix
degradation, indicating that Notch1 may be involved in cell
invasion.
Discussion
The Notch signaling pathway may be aberrantly
activated, and contributes to the development, invasion and
metastasis of a wide variety of human cancers, including cervical,
lung, colon, head and neck, renal carcinoma, acute myeloid, Hodgkin
and large-cell lymphomas and pancreatic cancer (19,20).
Notch1 has also been reported to be involved in the metastasis of
tumors, including osteosarcoma, breast cancer, melanoma and
prostate cancer (21–23). In spite of these findings, the
involvement of Notch1, in particular aberrantly-activated Notch1
signaling in prostate cancer cell metastasis, requires
verification. In the present study, the activity of Notch-1 was
investigated by shRNA-knockdown in human PCa LNCaP cells. Notch1
was revealed to be overexpressed in LNCaP, PC-3 and DU 145 cells
compared with RWPE-1 cells, and cleaved-Notch1 was expressed in
LNCaP, PC-3 and DU 145 cells, but not RWPE-1 cells. These results
were consistent with the findings of Bin Hafeez et al
(24), indicating that Notch
signaling is involved in prostate cancer development. It was also
observed that knockdown of Notch1 by shRNA in LNCaP cells markedly
decreased cell invasion through Matrigel, which mimicked the in
vivo extracellular matrix, and cell proliferation was also
inhibited 48 h post-transfection. The reduction in migration,
invasion and proliferation in PCa cells has been demonstrated to be
associated with Notch1-knockdown by RNA interference (24–26). The
present results verified the involvement of Notch1 in prostate
cancer cell metastasis.
The present study revealed that the expression of
MTA1 decreased significantly following RNA interference by Notch1
shRNA. MTA1 was initially identified in breast cancer (27), and was classified as a
metastasis-associated protein. It exists primarily in the nucleus
as a constituent part of the nucleosome remodeling and histone
deacetylation complex (28).
Subsequently, MTA1 was also detected in the cytoplasm (29).
Xue et al (30)
reported that MTA1 exhibits a strong inhibitory activity for the
transcription of a variety of tumor suppressor genes. MTA1 is a
stress response protein (31).
Multiple tumors have been confirmed to be associated with MTA1,
including breast cancer and colorectal cancer (32). High expression levels of MTA1 may help
cells to migrate to more hospitable areas in order to survive in
adverse conditions, including hypoxia (33). An additional study has also indicated
that overexpression of MTA1 is consistent with a more advanced
tumor stage and increases the rate of metastasis (34). An increase of KISS-1, MKK4 and KAI1
was also observed in Notch1-knockdown cells, indicating that Notch1
may regulate the expression of MTA1, KISS-1, MKK4 and KAI1.
The present data demonstrated the involvement of
Notch1 in human PCa invasion and that knockdown of Notch1 inhibited
invasion of human PCa cells by downregulating the expression of
MTA1 and upregulating the expression of KISS-1, MKK4 and KAI1.
These findings indicated that targeting Notch1 may be a novel
therapeutic approach for the treatment of prostate cancer
metastasis.
Acknowledgements
The authors would like to thank Zhejiang Academy of
Medical Sciences (Hangzhou, China) for expert technical assistance.
This study was funded by the Natural Science Foundation of Zhejiang
Province (Y2111329, LY13H310004, LY17H050002), the Science and
Technology Plan Projects of Zhejiang Province (2014C37016), the
National Natural Science Foundation of China (81172072), the
Science Foundation for Distinguished Young Scholars of Zhejiang
(R2101405), the Chinese Medicine Science and Technology Plan
Projects of Zhejiang Province (2016ZB099, 2013ZA107, 2011ZB099),
the Medicine and Health Science and Technology Plan Projects of
Zhejiang Province (2011KYB066, 2015KYB295), the Science and
Technology Plan Projects of Hangzhou (2017A05, 20110833B05,
20110733Q12), the Major Science and Technology Innovation Project
of Hangzhou (20112313A01) and the Collaborative Innovation Projects
of Science and Technology Department of Zhejiang Province
(2014F50014).
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2015. CA Cancer J Clin. 65:5–29. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bubendorf L, Schöpfer A, Wagner U, Sauter
G, Moch H, Willi N, Gasser TC and Mihatsch MJ: Metastatic patterns
of prostate cancer: An autopsy study of 1,589 patients. Hum Pathol.
31:578–583. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Lawton A, Sudakoff G, Dezelan LC and Davis
N: Presentation, treatment, and outcomes of dural metastases in men
with metastatic castrate-resistant prostate cancer: A case series.
J Palliat Med. 13:1125–1129. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
B R and C GP: Path to facilitate the
prediction of functional amino acid substitutions in red blood cell
disorders - a computational approach. PLoS One. 6:e246072011.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Carvalho FL, Simons BW, Eberhart CG and
Berman DM: Notch signaling in prostate cancer: A moving target.
Prostate. 74:933–945. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Vinson KE, George DC, Fender AW, Bertrand
FE and Sigounas G: The notch pathway in colorectal cancer. Int J
Cancer. 138:1835–1842. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Eliasz S, Liang S, Chen Y, De Marco MA,
Machek O, Skucha S, Miele L and Bocchetta M: Notch-1 stimulates
survival of lung adenocarcinoma cells during hypoxia by activating
the IGF-1R pathway. Oncogene. 29:2488–2498. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Mumm JS and Kopan R: Notch signaling: From
the outside in. Dev Biol. 228:151–165. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wang Z, Li Y, Banerjee S and Sarkar FH:
Exploitation of the Notch signaling pathway as a novel target for
cancer therapy. Anticancer Res. 28:3621–3630. 2008.PubMed/NCBI
|
|
10
|
Talora C, Campese AF, Bellavia D, Felli
MP, Vacca A, Gulino A and Screpanti I: Notch signaling and
diseases: An evolutionary journey from a simple beginning to
complex outcomes. Biochim Biophys Acta. 1782:489–497. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kopan R and Ilagan MX: The canonical Notch
signaling pathway: Unfolding the activation mechanism. Cell.
137:216–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Hori K, Sen A and Artavanis-Tsakonas S:
Notch signaling at a glance. J Cell Sci. 126:2135–2140. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Hudson BD, Kulp KS and Loots GG: Prostate
cancer invasion and metastasis: Insights from mining genomic data.
Brief Funct Genomics. 12:397–410. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Balint K, Xiao M, Pinnix CC, Soma A, Veres
I, Juhasz I, Brown EJ, Capobianco AJ, Herlyn M and Liu ZJ:
Activation of Notch1 signaling is required for
beta-catenin-mediated human primary melanoma progression. J Clin
Invest. 115:3166–3176. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yeh TS, Wu CW, Hsu KW, Liao WJ, Yang MC,
Li AF, Wang AM, Kuo ML and Chi CW: The activated Notch1 signal
pathway is associated with gastric cancer progression through
cyclooxygenase-2. Cancer Res. 69:5039–5048. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Negri FV, Crafa P, Pedrazzi G, Bozzetti C,
Lagrasta C, Gardini G, Tamagnini I, Bisagni A, Azzoni C, Bottarelli
L, et al: Strong Notch activation hinders bevacizumab efficacy in
advanced colorectal cancer. Future Oncol. 11:3167–3174. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ponnurangam S, Dandawate PR, Dhar A,
Tawfik OW, Parab RR, Mishra PD, Ranadive P, Sharma R, Mahajan G,
Umar S, et al: Quinomycin A targets Notch signaling pathway in
pancreatic cancer stem cells. Oncotarget. 7:3217–3232. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Lefort K, Ostano P, Mello-Grand M, Calpini
V, Scatolini M, Farsetti A, Dotto GP and Chiorino G: Dual tumor
suppressing and promoting function of Notch1 signaling in human
prostate cancer. Oncotarget. 7:48011–48026. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Stylianou S, Clarke RB and Brennan K:
Aberrant activation of notch signaling in human breast cancer.
Cancer Res. 66:1517–1525. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Agrawal N, Frederick MJ, Pickering CR,
Bettegowda C, Chang K, Li RJ, Fakhry C, Xie TX, Zhang J, Wang J, et
al: Exome sequencing of head and neck squamous cell carcinoma
reveals inactivating mutations in NOTCH1. Science. 333:1154–1157.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Leong KG, Niessen K, Kulic I, Raouf A,
Eaves C, Pollet I and Karsan A: Jagged1-mediated Notch activation
induces epithelial-to-mesenchymal transition through Slug-induced
repression of E-cadherin. J Exp Med. 204:2935–2948. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Chen J, Imanaka N, Chen J and Griffin JD:
Hypoxia potentiates Notch signaling in breast cancer leading to
decreased E-cadherin expression and increased cell migration and
invasion. Br J Cancer. 102:351–360. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Santagata S, Demichelis F, Riva A,
Varambally S, Hofer MD, Kutok JL, Kim R, Tang J, Montie JE,
Chinnaiyan AM, et al: JAGGED1 expression is associated with
prostate cancer metastasis and recurrence. Cancer Res.
64:6854–6857. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Bin Hafeez B, Adhami VM, Asim M, Siddiqui
IA, Bhat KM, Zhong W, Saleem M, Din M, Setaluri V and Mukhtar H:
Targeted knockdown of Notch1 inhibits invasion of human prostate
cancer cells concomitant with inhibition of matrix
metalloproteinase-9 and urokinase plasminogen activator. Clin
Cancer Res. 15:452–459. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhang Y, Wang Z, Ahmed F, Banerjee S, Li Y
and Sarkar FH: Down-regulation of Jagged-1 induces cell growth
inhibition and S phase arrest in prostate cancer cells. Int J
Cancer. 119:2071–2077. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Shou J, Ross S, Koeppen H, de Sauvage FJ
and Gao WQ: Dynamics of notch expression during murine prostate
development and tumorigenesis. Cancer Res. 61:7291–7297.
2001.PubMed/NCBI
|
|
27
|
Toh Y, Pencil SD and Nicolson GL: A novel
candidate metastasis-associated gene, mta1, differentially
expressed in highly metastatic mammary adenocarcinoma cell lines.
cDNA cloning, expression, and protein analyses. J Biol Chem.
269:22958–22963. 1994.PubMed/NCBI
|
|
28
|
Toh Y and Nicolson GL: The role of the MTA
family and their encoded proteins in human cancers: Molecular
functions and clinical implications. Clin Exp Metastasis.
26:215–227. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Liu J, Xu D, Wang H, Zhang Y, Chang Y,
Zhang J, Wang J, Li C, Liu H, Zhao M, et al: The subcellular
distribution and function of MTA1 in cancer differentiation.
Oncotarget. 5:5153–5164. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Xue Y, Wong J, Moreno GT, Young MK, Côté J
and Wang W: NURD, a novel complex with both ATP-dependent
chromatin-remodeling and histone deacetylase activities. Mol Cell.
2:851–861. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Li J, Ye L, Sun PH, Satherley L, Hargest
R, Zhang Z and Jiang WG: MTA1 Is Up-regulated in colorectal cancer
and is inversely correlated with lymphatic metastasis. Cancer
Genomics Proteomics. 12:339–345. 2015.PubMed/NCBI
|
|
32
|
Kang HJ, Lee MH, Kang HL, Kim SH, Ahn JR,
Na H, Na TY, Kim YN, Seong JK and Lee MO: Differential regulation
of estrogen receptor α expression in breast cancer cells by
metastasis-associated protein 1. Cancer Res. 74:1484–1494. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Wang RA: MTA1-a stress response protein: A
master regulator of gene expression and cancer cell behavior.
Cancer Metastasis Rev. 33:1001–1009. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Luo H, Li H, Yao N, Hu L and He T:
Metastasis-associated protein 1 as a new prognostic marker for
solid tumors: A meta-analysis of cohort studies. Tumour Biol.
35:5823–5832. 2014. View Article : Google Scholar : PubMed/NCBI
|