Introduction
Osteosarcoma (OS) is a common cancerous bone tumor
most prevalent in children and young adults (1). Specifically, it is a histological form
of primary bone cancer derived from primitive transformed cells of
mesenchymal origin (2). Numerous
patients with OS also suffer from panic attacks and swelling of the
lower femur or area directly inferior to the knee, and these
symptoms are often exacerbated at night (3). The cause of OS is unknown; however, it
is suggested that this disease may be associated with several
factors including inheritance, bone dysplasia, germline p53
mutations and Rothmund-Thomson syndrome (4).
p53 is a tumor suppressor gene that regulates
the expression of apoptosis-associated genes when stimulated by
specific molecular signals (5).
p53 mutations have been revealed to be associated with the
development of OS (6,7). Luo et al (8) constructed a regulatory network of OS,
and further screened IL-6 and BCL2L1 as target genes
regulated by p53.
U2OS is a commonly utilized OS cell line. Various
chemotherapy drugs, including actinomycin D (ActD), doxorubicin
(DXR), Nutlin-3 and etoposide (Eto), have been widely used in OS
treatment. Among these drugs, ActD (9), DXR (10)
and Eto (11) exhibit direct effects
on DNA, inhibiting transcription and promoting apoptosis. However,
Nutlin-3 interacts with and disrupts mouse double minute 2 homolog
(MDM2), a negative regulator of p53. Inhibiting the interaction
between MDM2 and p53 results in an increase in activated p53 and
therefore apoptosis (12). In
addition, the four drugs can induce cell cycle arrest (13–15). The
cell-protective agent dimethyl sulfoxide (DMSO) has also been
revealed to affect p53 (16).
In order to investigate the response of p53 to the
various drugs in the U2OS cell line, p53 chromatin
immunoprecipitation combined with sequencing (ChIP-seq) and
microarray data of ActD, DXR, Nutlin-3 and Eto treatment were
downloaded for analysis of molecular mechanism.
Differentially-expressed genes (DEGs) were screened prior to
alignment analysis. Finally, the target genes were investigated for
the construction of regulatory networks and annotations were
processed.
Materials and methods
Data sources
The microarray datasets and p53 ChIP-seq datasets of
OS cell line U2OS treated with distinct drugs (17,18) were
acquired from the Gene Expression Omnibus (GEO; www.ncbi.nlm.nih.gov/geo) database. The U2OS cell
line was treated with various drugs, including DMSO, DXR, ActD,
Nutlin-3 and Eto (Table I).
 | Table I.p53 ChIP combined with sequencing
datasets. |
Table I.
p53 ChIP combined with sequencing
datasets.
| Author | Gene expression
omnibus sample | Description of U2OS
cells | (Refs) |
|---|
| Menendez et
al |
GSM1133482 | DMSO-treated
ChIP | (17) |
|
|
GSM1133483 | DMSO-treated
input |
|
|
|
GSM1133484 | DXR-treated
ChIP |
|
|
|
GSM1133485 | DXR-treated
input |
|
|
|
GSM1133486 | Nutlin-3-treated
ChIP |
|
|
|
GSM1133487 | Nutlin-3-treated
input |
|
|
|
GSM1133488 | No treatment
ChIP |
|
|
|
GSM1133489 | No treatment
input |
|
| Smeenk et
al | GSM545807 | ActD-treated
ChIP | (18) |
|
| GSM545808 | Etoposide-treated
ChIP |
|
Analytical methods
DEG analysis. The downloaded microarray
datasets of ActD and Eto were standardized. Compared with drug
treatment groups and the control U2OS cells without any drug
treatment, genes with |log2(fold-change)|>1 were
considered to be DEGs. Raw data from DXR, Nutlin-3 and DMSO
microarray datasets were processed by Affy analysis of the
Bioconductor 2.0 in R (http://www.bioconductor.org/packages/release/bioc/html/affy.html)
(19) with P<0.001 and
|log2(fold-change)|>1 considered to indicate
DEGs.
Alignment and annotation of gene
sequences
Bowtie 2 (version 2.0.0-beta5; http://bowtie-bio.sourceforge.net/bowtie2/index.shtml)
(20), a tool for aligning sequencing
reads to long reference sequences, was utilized for gene sequence
alignment between ChIP-seq and Human Genome hg19. Model-based
analysis of ChIP-Seq 2 was applied to identify peaks of
transcription factor p53-binding regions (21). The two procedures used the default
value as parameter. Peak annotations were processed by CisGenome
(version 2.0; http://www.biostat.jhsph.edu/~hji/cisgenome/)
(22), an integrated tool for tiling
array, ChIP-seq, genome and cis-regulatory element analysis.
Any genes presenting with a peak located between 2,000 bp upstream
and 1,000 bp downstream of the transcription start site was
considered to be a p53-binding target gene.
Screening and annotation of target
genes
The data gathered from ChIP-seq and expression
profile microarray were combined to further screen p53 target genes
in the U2OS cell line (promoter range, 2,000±500 bp; the remaining
parameters were set using the default values). Subsequently, a
p53-centered expression network was constructed. Finally, the
Database for Annotation, Visualization and Integrated Discovery
(http://david.niaid.nih.gov) (23), an analytical tool for extracting
biological information from large lists of genes, was used for
annotation of target genes.
Results
Target genes of p53 binding
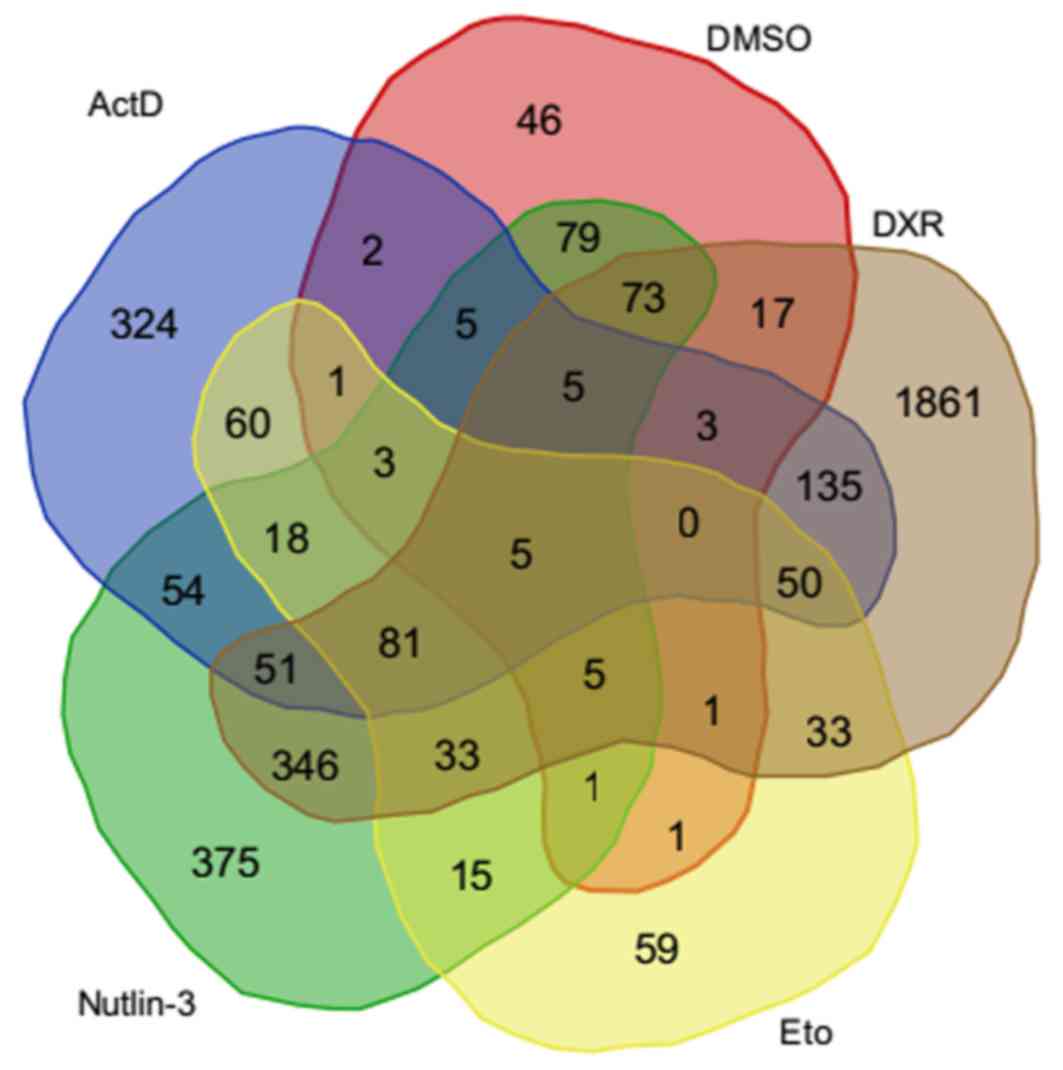
A total of 212 p53-binding peaks were identified in
the untreated group, whereas thousands of peaks were obtained in
the treated groups (Table II).
Similar numbers of binding sites were identified in the ActD, DXR,
Eto and DMSO treatment groups, respectively, with ~1,000 target
genes, whereas a total of 5,458 target genes were obtained in the
Nutlin-3 treatment group. There were 504 common target genes across
the five treatment groups (Fig. 1).
Moreover, these target genes were significantly enriched in GO
functions associated with positive regulation of apoptosis,
positive regulation of programmed cell death and positive
regulation of cell death (Table
III). Notably, programmed cell death can be divided into
several categories including type I (apoptosis) and type II
(autophagic death) (24), thus,
target genes enriched in positive regulation of apoptosis were
different from those enriched in positive regulation of programmed
cell death.
 | Table II.Microarray datasets. |
Table II.
Microarray datasets.
| Author | Gene expression
omnibus sample | Description of U2OS
cells | (Refs) |
|---|
| Menendez et
al |
GSM1131226 | No treatment repeat
1 | (17) |
|
|
GSM1131227 | No treatment repeat
2 |
|
|
|
GSM1131228 | No treatment repeat
3 |
|
|
|
GSM1131229 | DXR-treated repeat
1 |
|
|
|
GSM1131230 | DXR-treated repeat
2 |
|
|
|
GSM1131231 | DXR-treated repeat
3 |
|
|
|
GSM1131232 | DMSO-treated repeat
1 |
|
|
|
GSM1131233 | DMSO-treated repeat
2 |
|
|
|
GSM1131234 | DMSO-treated repeat
3 |
|
|
|
GSM1131235 | Nutlin-3-treated
repeat 1 |
|
|
|
GSM1131236 | Nutlin-3-treated
repeat 2 |
|
|
|
GSM1131237 | Nutlin-3-treated
repeat 3 |
|
| Smeenk et
al | GSM552391 | Control ActD repeat
1 | (18) |
|
| GSM552392 | Control ActD repeat
2 |
|
|
| GSM552393 | ActD repeat 1 |
|
|
| GSM552394 | ActD repeat 2 |
|
|
| GSM552395 | Control Eto repeat
1 |
|
|
| GSM552396 | Control Eto repeat
2 |
|
|
| GSM552397 | Eto repeat 1 |
|
|
| GSM552398 | Eto repeat 2 |
|
 | Table III.Gene ontology analysis of p53 target
genes. |
Table III.
Gene ontology analysis of p53 target
genes.
| Gene ontology
number | Role | n | Genes | False discovery
rate, ×10−4 |
|---|
| 0043065 | Positive regulation
of apoptosis | 32 | ZAK, IL19,
RPS27L, RRM2B, BCL2L1, SRC, ZC3H8, GPX1, AEN,
FAS, PHLDA3, FGD3, ARHGEF3, PTPRF, HTT, PRKCE, VAV2,
TNFSF8, PLEKHF1, NOTCH2, CDKN1A, TNFRSF10B, NUPR1,
BBC3, LYST, BAX, FAF1, ABL1, DCUN1D3, APBB2, NGF,
KALRN | 1.92 |
| 0043068 | Positive regulation
of programmed cell death | 32 | ZAK, IL19,
RPS27L, RRM2B, BCL2L1, SRC, ZC3H8, GPX1, AEN, FAS,
PHLDA3, FGD3, ARHGEF3, PTPRF, HTT, PRKCE, VAV2,
TNFSF8, PLEKHF1, NOTCH2, CDKN1A, TNFRSF10B, NUPR1,
BBC3, LYST, BAX, FAF1, ABL1, DCUN1D3, APBB2,
NGF, KALRN | 2.24 |
| 0010942 | Positive regulation
of cell death | 32 | ZAK, IL19,
RPS27L, RRM2B, BCL2L1, SRC, ZC3H8, GPX1, AEN, FAS,
PHLDA3, FGD3, ARHGEF3, PTPRF, HTT, PRKCE, VAV2,
TNFSF8, PLEKHF1, NOTCH2, CDKN1A, TNFRSF10B, NUPR1,
BBC3, LYST, BAX, FAF1, ABL1, DCUN1D3, APBB2, NGF,
KALRN | 2.48 |
| 0006974 | Response to DNA
damage stimulus | 28 | RAD51C, ZAK,
RPS27L, RRM2B, SESN1, TRIAP1, RAD51L1, AEN, NSMCE2,
PHLDA3, FANCC, POLH, WRN, FOXN3, CDKN1A,
ATXN3, RFC3, EYA2, NUPR1, BTG2, BBC3, BAX,
DDB2, BRE, PCNA, ABL1, GADD45A, REV3L | 11.98 |
| 0033554 | Cellular response
to stress | 35 | RAD51C, ZAK,
ADORA2B, RTN4RL1, RPS27L, RRM2B, SESN1, GPX1, TRIAP1,
RAD51L1, AEN, TPO, NSMCE2, TRPV4, PHLDA3, FANCC,
POLH, WRN, MAPK10, FOXN3, RFC3, CDKN1A, ATXN3,
EYA2, NUPR1, BTG2, BBC3, BAX, ATP2A1,
DDB2, BRE, PCNA, ABL1, GADD45A, REV3L | 32.40 |
| 0006917 | Induction of
apoptosis | 24 | ARHGEF3, HTT,
IL19, RPS27L, RRM2B, VAV2, PRKCE, TNFSF8, PLEKHF1,
NOTCH2, GPX1, CDKN1A, TNFRSF10B, NUPR1, BBC3, AEN,
BAX, LYST, FAS, ABL1, PHLDA3, FGD3, NGF, KALRN | 93.17 |
| 0012502 | Induction of
programmed cell death | 24 | ARHGEF3, HTT,
IL19, RPS27L, RRM2B, VAV2, PRKCE, TNFSF8, PLEKHF1,
NOTCH2, GPX1, CDKN1A, TNFRSF10B, NUPR1, BBC3, AEN,
BAX, LYST, FAS, ABL1, PHLDA3, FGD3, NGF, KALRN | 98.13 |
Distinct responses to various
drugs
A total of five DEGs including AREG, LPP, ATF3,
FAM198B and HAPLN1 were revealed across each of the five
treatment groups (Fig. 1).
Furthermore, a total of 86 common DEGs were obtained from the ActD,
DXR, Eto and Nutlin-3 treatment groups, which were classified as
Gene Ontology (GO) terms including p53 signaling pathway, cell
adhesion and biological adhesion (Table
IV). Several common DEGs including MDM2, TP53I3, RRM2B,
FAS and SESN1 identified in these four treatment groups
were also target genes for p53 binding (Table III).
 | Table IV.Gene ontology and KEGG enrichment
analysis of differentially expressed genes in doxorubicin,
Nutlin-3, actinomycin D and etoposide treatment groups. |
Table IV.
Gene ontology and KEGG enrichment
analysis of differentially expressed genes in doxorubicin,
Nutlin-3, actinomycin D and etoposide treatment groups.
| Gene ontology/KEGG
number | Role | n | Genes |
|---|
| hsa04115 | p53 signaling
pathway | 5 | TP53I3, MDM2,
RRM2B, FAS, SESN1 |
| 0007155 | Cell adhesion | 11 | HAPLN1, PVRL4,
COL17A1, LPP, PKP4, CYFIP2, NINJ1, KITLG,
SLAMF7, NEGR1, FEZ1 |
| 0022610 | Biological
adhesion | 11 | HAPLN1, PVRL4,
COL17A1, LPP, PKP4, CYFIP2, NINJ1, KITLG, SLAMF7,
NEGR1, FEZ1 |
| 0042981 | Regulation of
apoptosis | 11 | TRIAP1, TP53I3,
NUPR1, BTG2, BTG1, RRM2B, FAS, SLAMF7, NEFL, ANXA4,
TP53INP1 |
| 0043067 | Regulation of
programmed cell death | 11 | TRIAP1, TP53I3,
NUPR1, BTG2, BTG1, RRM2B, FAS, SLAMF7, NEFL, ANXA4,
TP53INP1 |
| 0010941 | Regulation of cell
death | 11 | TRIAP1, TP53I3,
NUPR1, BTG2, BTG1, RRM2B, FAS, SLAMF7, NEFL,
ANXA4, TP53INP1 |
| 0008083 | Growth factor
activity | 5 | TGFA, KITLG,
ESM1, AREG, GDF15 |
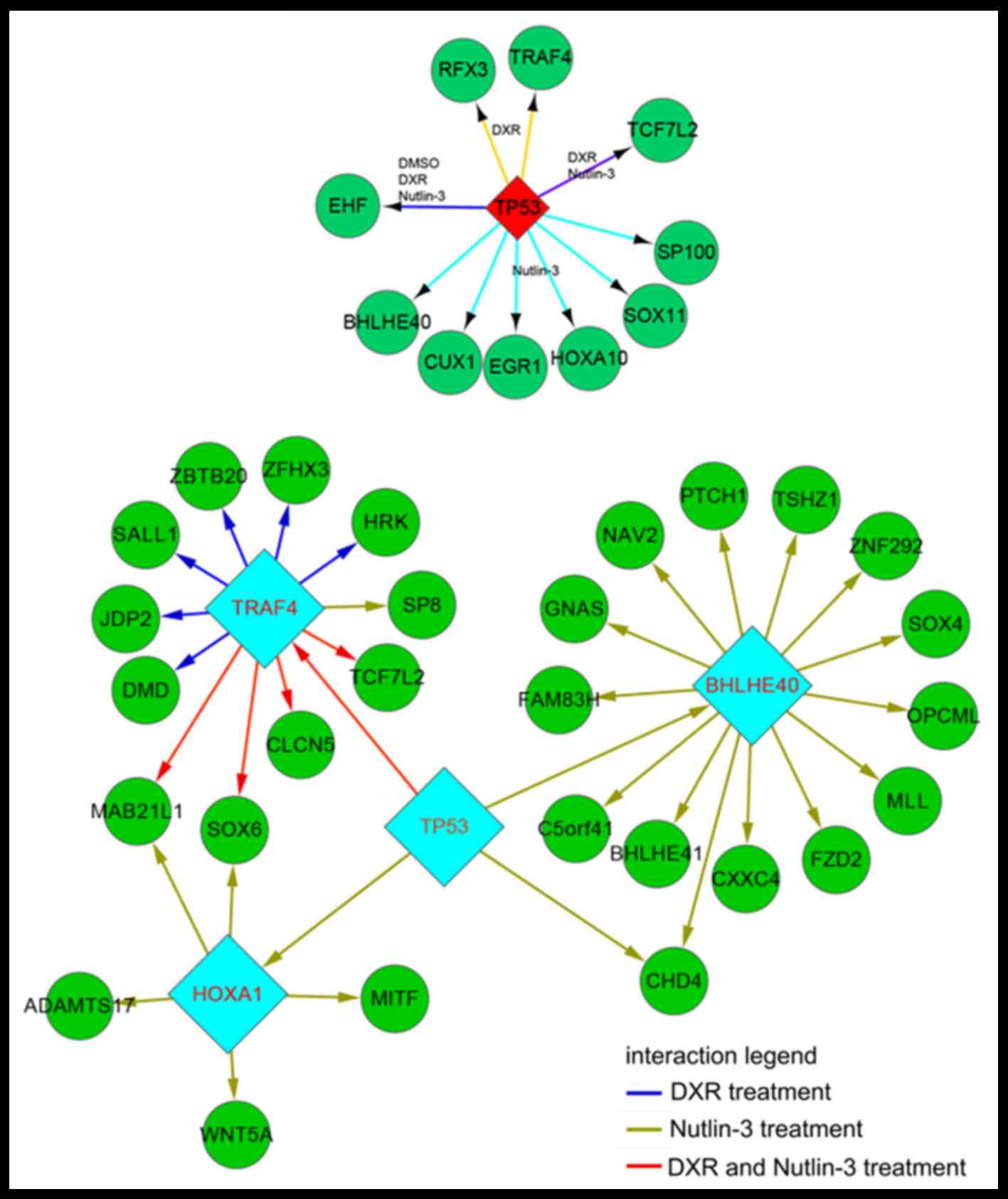
p53 indirectly regulates downstream
genes through other key genes
p53 was able to activate downstream hub genes
following a number of drug treatments (Fig. 2). For example, p53 regulated various
genes including EHF, HOXA10 and BHLHE40 in the
Nutlin-3 treatment group, whereas p53 regulated EHF, RFX3,
TRAF40 and TCF7L2 in the DXR treatment group.
Additionally, p53 was able to indirectly regulate further genes
through TRAF4, BHLHE40 and HOXA10 hub genes (Fig. 2).
Discussion
Owing to systemic chemotherapy, long-term outcomes
for patients with OS have improved; however, subsequent progress
required further research (2). In the
present study, a total of five DEGs were revealed across all five
treatment groups including AREG, LPP, ATF3, FAM198B and
HAPLN1. Additionally, a total of 86 common DEGs were
obtained in each of the ActD, DXR, Eto and Nutlin-3 treatment
groups, certain of which were identified as being associated with
the p53 signaling pathway. Following treatment with various drugs,
p53 was identified to be able to activate downstream hub genes
including TRAF4, BHLHE40 and HOXA10 which was, in
turn, able to affect more genes.
DMSO is a cell-protective agent with limited genetic
effects. Of the 86 common DEGs obtained in the four other treatment
groups (ActD, DXR, Eto and Nutlin-3), only five were affected by
DMSO. In the p53 signaling pathway, DEGs including MDM2,
TP53I3 and RRM2B were enriched. MDM2 encodes a
nuclear-localized E3 ubiquitin ligase, targets p53 and
further promotes tumor formation (25). Soft tissue sarcoma and malignant
fibrous histiocytoma are common diseases associated with
MDM2 (26). E3
ubiquitin-protein ligase is able to lead to the degradation of p53
by the proteasome and further inhibits cell cycle arrest and
apoptosis by binding the transcriptional activation domain
(27). In addition, TP53I3 was
also differentially expressed in the non-DMSO treatment groups.
TP53I3 is a protein-coding gene which encodes enzymes
involved in cellular responses to irradiation and oxidative stress
(28). This gene is considered to be
induced by p53 and involved in p53-mediated cell death (29). TP53I3 is transcriptionally
activated by p53 through interacting with downstream
pentanucleotide microsatellite sequences, and is associated with
the number of pentanucleotide repeats. Furthermore, the
microsatellite polymorphism is closely associated with the
differential susceptibility to cancer (30). Additionally, RRM2B encodes the
small subunit of p53-incucible ribonucleoside reductase which
catalyzes the conversion of ribonucleoside into deoxyribonucleoside
diphosphates (31). This gene serves
a crucial role in cell survival through repairing DNA in a
p53-dependent manner (31). In the
process of cell cycle arrest, RRM2B also participates in DNA
repair by supplying deoxyribonucleotides (32). Therefore, DEGs including MDM2,
TP53I3 and RRM2B may be target genes for p53
binding.
In addition to the aforementioned genes, certain
downstream genes of p53 may also be affected by drugs. The present
study revealed that p53 was able to regulate EHF which may
in turn regulate further genes in the DMSO, DXR and Nutlin-3
treatment groups. EHF encodes a protein that is a member of
the E26 transformation-specific transcription factor subfamily
(33). The encoded protein may
participate in carcinogenesis and epithelial differentiation as a
transcriptional repressor (34). In
addition, a previous study has demonstrated that EHF may
perform roles in molecular processes including sequence-specific
DNA-binding transcription factor activity and sequence-specific DNA
binding (35). Additionally, in the
DXR treatment group, p53 was able to regulate hub genes including
RFX3 to further regulate more genes. RFX3, a member
of the regulatory factor X gene family, encodes a transcriptional
activator protein (36). This protein
is able to bind to DNA with other RFX family members
(37). As with EHF, GO
annotations associated with RFK3 exhibited sequence-specific
DNA-binding transcription factor activity (38). Subsequently, p53 was able to
indirectly regulate genes through several hub genes including
EHF and RFX in the U2OS cells treated with a number
of drugs.
The results of the present study indicates that p53
is able to directly regulate target genes including MDM2,
TP53I3 and RRM2B or indirectly regulate more genes
through several hub genes including EHF and RFX as
demonstrated using various treatments of U2OS cells. Furthermore,
p53 may be involved in distinct molecular processes regulated by
various drug treatments. However, further experimental analysis is
required to confirm these results.
References
|
1
|
Burke ME, Albritton K and Marina N:
Challenges in the recruitment of adolescents and young adults to
cancer clinical trials. Cancer. 110:2385–2393. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Luetke A, Meyers PA, Lewis I and Juergens
H: Osteosarcoma treatment-where do we stand? A state of the art
review. Cancer Treat Rev. 40:523–532. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kere J: Neuropeptide S receptor 1: An
asthma susceptibility geneAllergy Frontiers: Future Perspectives.
Springer; pp. 191–205. 2010, View Article : Google Scholar
|
|
4
|
Ahmed H, Salama A, Salem SE and Bahnassy
AA: A case of synchronous double primary breast carcinoma and
osteosarcoma: Mismatch repair genes mutations as a possible cause
for multiple early onset malignant tumors. Am J Case Rep.
13:218–223. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Vogelstein B, Lane D and Levine AJ:
Surfing the p53 network. Nature. 408:307–310. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Mulligan LM, Matlashewski GJ, Scrable HJ
and Cavenee WK: Mechanisms of p53 loss in human sarcomas. Proc Natl
Acad Sci USA. 87:5863–5867. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Toguchida J, Yamaguchi T, Dayton SH,
Beauchamp RL, Herrera GE, Ishizaki K, Yamamuro T, Meyers PA, Little
JB, Sasaki MS, et al: Prevalence and spectrum of germline mutations
of the p53 gene among patients with sarcoma. N Engl J Med.
326:1301–1308. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Luo Y, Deng Z and Chen J: Pivotal
regulatory network and genes in osteosarcoma. Arch Med Sci.
9:569–575. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Sobell HM: Actinomycin and DNA
transcription. Proc Natl Acad Sci USA. 82:5328–5331. 1985.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Fornari FA, Randolph JK, Yalowich JC,
Ritke MK and Gewirtz DA: Interference by doxorubicin with DNA
unwinding in MCF-7 breast tumor cells. Mol Pharmacol. 45:649–656.
1994.PubMed/NCBI
|
|
11
|
Hande KR: Etoposide: Four decades of
development of a topoisomerase II inhibitor. Eur J Cancer.
34:1514–1521. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Shinohara T and Uesugi M: In-vivo
activation of the p53 pathway by small-molecule antagonists of
MDM2. Tanpakushitsu Kakusan Koso. 52 (13 Suppl):S1816–S1817.
2007.
|
|
13
|
Miyachi M, Kakazu N, Yagyu S, Katsumi Y,
Tsubai-Shimizu S, Kikuchi K, Tsuchiya K, Iehara T and Hosoi H:
Restoration of p53 pathway by nutlin-3 induces cell cycle arrest
and apoptosis in human rhabdomyosarcoma cells. Clin Cancer Res.
15:4077–4084. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ling YH, el-Naggar AK, Priebe W and
Perez-Soler R: Cell cycle-dependent cytotoxicity, G2/M phase
arrest, and disruption of p34cdc2/cyclin B1 activity induced by
doxorubicin in synchronized P388 cells. Mol Pharmacol. 49:832–841.
1996.PubMed/NCBI
|
|
15
|
Xu H and Krystal GW: Actinomycin D
decreases Mcl-1 expression and acts synergistically with ABT-737
against small cell lung cancer cell lines. Clin Cancer Res.
16:4392–4400. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hsiao M, Low J, Dorn E, Ku D, Pattengale
P, Yeargin J and Haas M: Gain-of-function mutations of the p53 gene
induce lymphohematopoietic metastatic potential and tissue
invasiveness. Am J Pathol. 145:7021994.PubMed/NCBI
|
|
17
|
Menendez D, Nguyen TA, Freudenberg JM,
Mathew VJ, Anderson CW, Jothi R and Resnick MA: Diverse stresses
dramatically alter genome-wide p53-binding and transactivation
landscape in human cancer cells. Nucleic Acids Res. 41:7286–7301.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Smeenk L, van Heeringen SJ, Koeppel M,
Gilbert B, Janssen-Megens E, Stunnenberg HG and Lohrum M: Role of
p53 serine 46 in p53 target gene regulation. PLoS One.
6:e175742011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Gautier L, Cope L, Bolstad BM and Irizarry
RA: affy-analysis of Affymetrix GeneChip data at the probe level.
Bioinformatics. 20:307–315. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Langmead B and Salzberg SL: Fast
gapped-read alignment with Bowtie 2. Nat Methods. 9:357–359. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhang Y, Liu T, Meyer CA, Eeckhoute J,
Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W and
Liu XS: Model-based analysis of ChIP-Seq (MACS). Genome Biol.
9:R1372008. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ji H, Jiang H, Ma W, Johnson DS, Myers RM
and Wong WH: An integrated software system for analyzing ChIP-chip
and ChIP-seq data. Nat Biotechnol. 26:1293–1300. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Huang DW, Sherman BT, Tan Q, Kir J, Liu D,
Bryant D, Guo Y, Stephens R, Baseler MW, Lane HC and Lempicki RA:
DAVID bioinformatics resources: Expanded annotation database and
novel algorithms to better extract biology from large gene lists.
Nucleic Acids Res. 35:W169–W175. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Shimizu S, Kanaseki T, Mizushima N, Mizuta
T, Arakawa-Kobayashi S, Thompson CB and Tsujimoto Y: Role of Bcl-2
family proteins in a non-apoptotic programmed cell death dependent
on autophagy genes. Nat Cell Biol. 6:1221–1228. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kim Y, Starostina NG and Kipreos ET: The
CRL4Cdt2 ubiquitin ligase targets the degradation of p21Cip1 to
control replication licensing. Genes Dev. 22:2507–2519. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Dujardin F, Binh MB, Bouvier C,
Gomez-Brouchet A, Larousserie F, Muret Ad, Louis-Brennetot C,
Aurias A, Coindre JM, Guillou L, et al: MDM2 and CDK4
immunohistochemistry is a valuable tool in the differential
diagnosis of low-grade osteosarcomas and other primary
fibro-osseous lesions of the bone. Mod Pathol. 24:624–637. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Lukin DJ, Carvajal LA, Liu WJ,
Resnick-Silverman L and Manfredi JJ: p53 promotes cell survival due
to the reversibility of its cell cycle checkpoints. Mol Cancer Res.
13:16–28. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Lee YS, Oh JH, Yoon S, Kwon MS, Song CW,
Kim KH, Cho MJ, Mollah ML, Je YJ, Kim YD, et al: Differential gene
expression profiles of radioresistant non-small-cell lung cancer
cell lines established by fractionated irradiation: Tumor protein
p53-inducible protein 3 confers sensitivity to ionizing radiation.
Int J Radiat Oncol Biol Phys. 77:858–866. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Voltan R, Secchiero P, Corallini F and
Zauli G: Selective induction of TP53I3/p53-inducible gene 3 (PIG3)
in myeloid leukemic cells, but not in normal cells, by Nutlin-3.
Mol Carcinog. 53:498–504. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
30
|
Qin X, Zhang S, Li B, Liu XD, He XP, Shang
ZF, Xu QZ, Zhao ZQ, Ye QN and Zhao PK: p53-dependent upregulation
of PIG3 transcription by γ-ray irradiation and its interaction with
KAP1 in responding to DNA damage. Chin Sci Bull. 56:3162–3171.
2011. View Article : Google Scholar
|
|
31
|
Bourdon A, Minai L, Serre V, Jais JP,
Sarzi E, Aubert S, Chrétien D, de Lonlay P, Paquis-Flucklinger V,
Arakawa H, et al: Mutation of RRM2B, encoding p53-controlled
ribonucleotide reductase (p53R2), causes severe mitochondrial DNA
depletion. Nat Genet. 39:776–780. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Kimura T, Takeda S, Sagiya Y, Gotoh M,
Nakamura Y and Arakawa H: Impaired function of p53R2 in Rrm2b-null
mice causes severe renal failure through attenuation of dNTP pools.
Nat Genet. 34:440–445. 2003. View
Article : Google Scholar : PubMed/NCBI
|
|
33
|
Kas K, Finger E, Grall F, Gu X, Akbarali
Y, Boltax J, Weiss A, Oettgen P, Kapeller R and Libermann TA:
ESE-3, a novel member of an epithelium-specific ets transcription
factor subfamily, demonstrates different target gene specificity
from ESE-1. J Biol Chem. 275:2986–2998. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Tugores A, Le J, Sorokina I, Snijders AJ,
Duyao M, Reddy PS, Carlee L, Ronshaugen M, Mushegian A, Watanaskul
T, et al: The epithelium-specific ETS protein EHF/ESE-3 is a
context-dependent transcriptional repressor downstream of MAPK
signaling cascades. J Biol Chem. 276:20397–20406. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Jolma A, Kivioja T, Toivonen J, Cheng L,
Wei G, Enge M, Taipale M, Vaquerizas JM, Yan J, Sillanpää MJ, et
al: Multiplexed massively parallel SELEX for characterization of
human transcription factor binding specificities. Genome Res.
20:861–873. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Maijgren S, Sur I, Nilsson M and Toftgård
R: Involvement of RFX proteins in transcriptional activation from a
Ras-responsive enhancer element. Arch Dermatol Res. 295:482–489.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Sengupta P, Xu Y, Wang L, Widom R and
Smith BD: Collagen alpha1(I) gene (COL1A1) is repressed by RFX
family. J Biol Chem. 280:21004–21014. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Badis G, Berger MF, Philippakis AA,
Talukder S, Gehrke AR, Jaeger SA, Chan ET, Metzler G, Vedenko A,
Chen X, et al: Diversity and complexity in DNA recognition by
transcription factors. Science. 324:1720–1723. 2009. View Article : Google Scholar : PubMed/NCBI
|