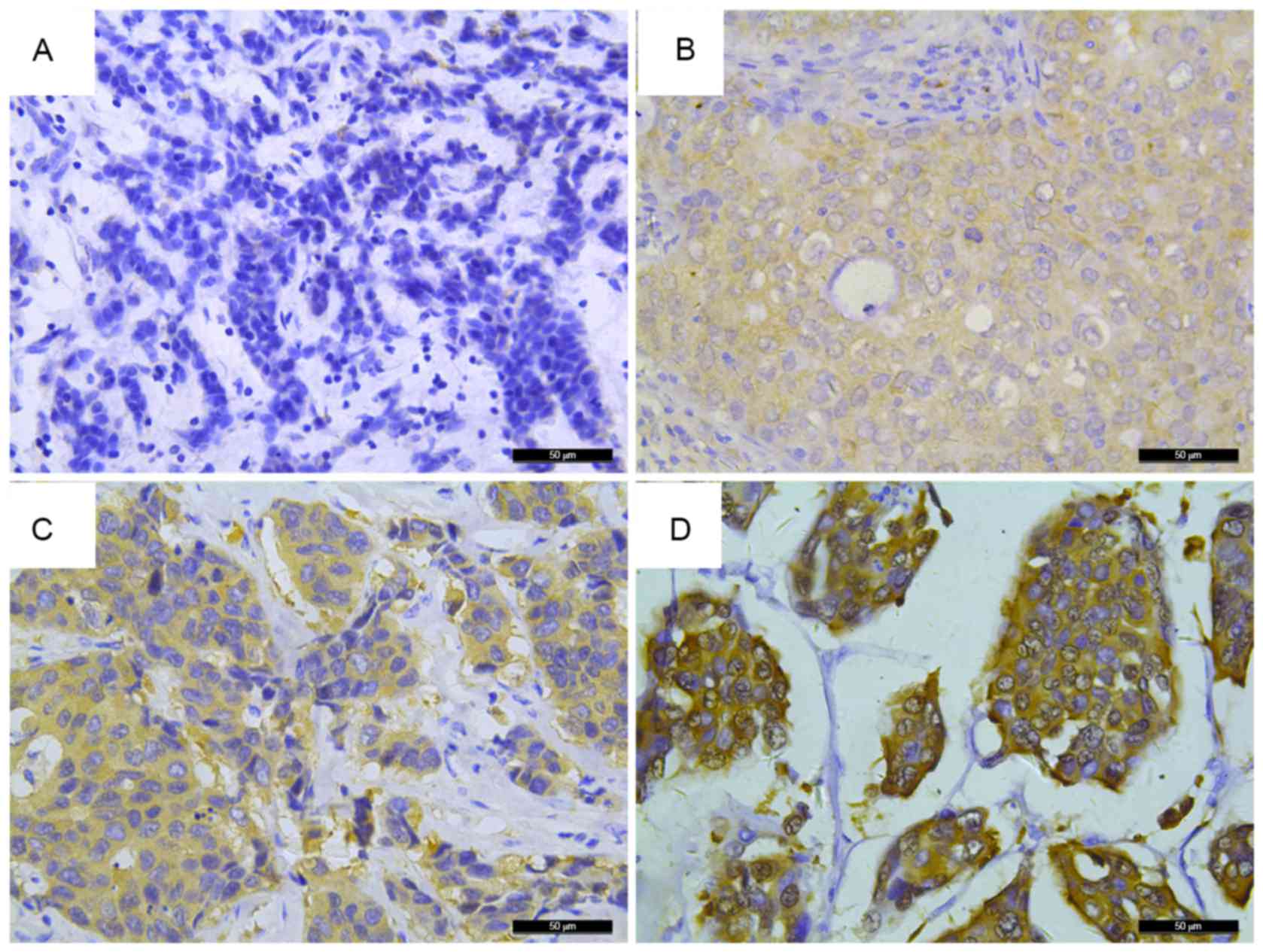
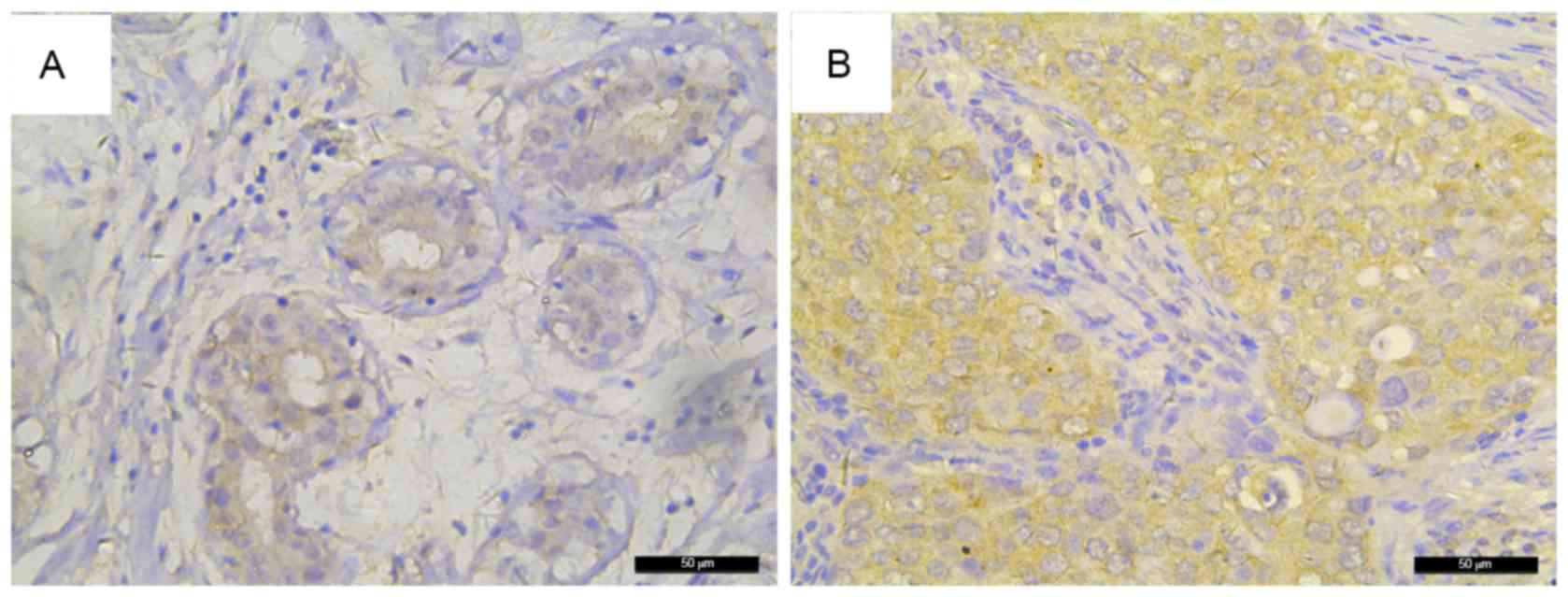
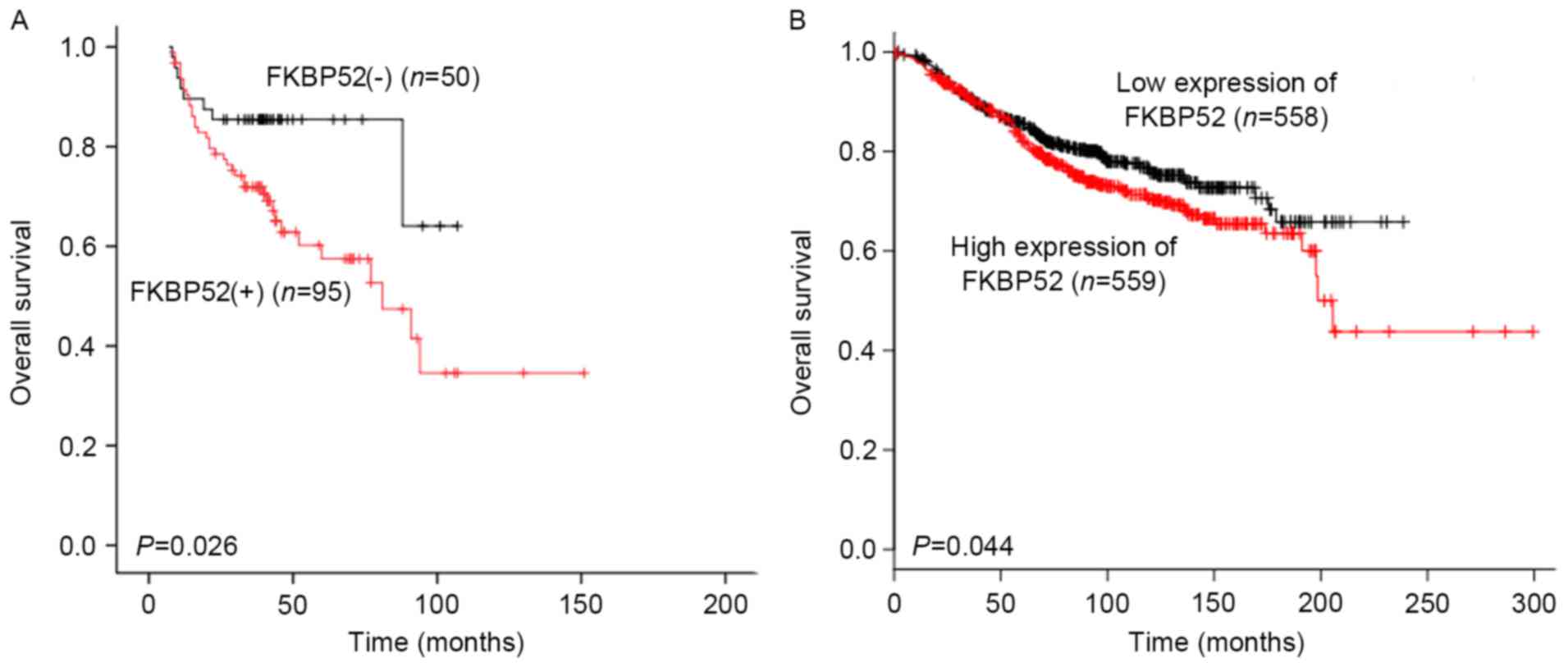
|
1
|
Siekierka JJ, Hung SH, Poe M, Lin CS and
Sigal NH: A cytosolic binding protein for the immunosuppressant
FK506 has peptidyl-prolyl isomerase activity but is distinct from
cyclophilin. Nature. 341:755–757. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Theuerkorn M, Fischer G and
Schiene-Fischer C: Prolyl cis/trans isomerase signalling pathways
in cancer. Curr Opin Pharmacol. 11:281–287. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Romano MF: FKBPs: Opportunistic modifiers
or active players in cancer? Curr Opin Pharmacol. 11:279–280. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Cioffi DL, Hubler TR and Scammell JG:
Organization and function of the FKBP52 and FKBP51 genes. Curr Opin
Pharmacol. 11:308–313. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Riggs DL, Roberts PJ, Chirillo SC,
Cheung-Flynn J, Prapapanich V, Ratajczak T, Gaber R, Picard D and
Smith DF: The Hsp90-binding peptidylprolyl isomerase FKBP52
potentiates glucocorticoid signaling in vivo. EMBO J. 22:1158–1167.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Pirkl F and Buchner J: Functional analysis
of the Hsp90-associated human peptidyl prolyl cis/trans isomerases
FKBP51, FKBP52 and Cyp40. J Mol Biol. 308:795–806. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Davies TH and Sánchez ER: FKBP52. Int J
Biochem Cell Biol. 37:42–47. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Galigniana MD, Harrell JM, O'Hagen HM,
Ljungman M and Pratt WB: Hsp90-binding immunophilins link p53 to
dynein during p53 transport to the nucleus. J Biol Chem.
279:22483–22489. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Teiten MH, Gaigneaux A, Chateauvieux S,
Billing AM, Planchon S, Fack F, Renaut J, Mack F, Muller CP, Dicato
M and Diederich M: Identification of differentially expressed
proteins in curcumin-treated prostate cancer cell lines. OMICS.
16:289–300. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lin JF, Xu J, Tian HY, Gao X, Chen QX, Gu
Q, Xu GJ, Song JD and Zhao FK: Identification of candidate prostate
cancer biomarkers in prostate needle biopsy specimens using
proteomic analysis. Int J Cancer. 121:2596–2605. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Liu Y, Li C, Xing Z, Yuan X, Wu Y, Xu M,
Tu K, Li Q, Wu C, Zhao M and Zeng R: Proteomic mining in the
dysplastic liver of WHV/c-myc mice-insights and indicators for
early hepatocarcinogenesis. FEBS J. 277:4039–4053. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Guerrero-Preston R, Hadar T, Ostrow KL,
Soudry E, Echenique M, Ili-Gangas C, Pérez G, Perez J,
Brebi-Mieville P, Deschamps J, et al: Differential promoter
methylation of kinesin family member 1a in plasma is associated
with breast cancer and DNA repair capacity. Oncol Rep. 32:505–512.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ostrow KL, Park HL, Hoque MO, Kim MS, Liu
J, Argani P, Westra W, Van Criekinge W and Sidransky D:
Pharmacologic unmasking of epigenetically silenced genes in breast
cancer. Clin Cancer Res. 15:1184–1191. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ward BK, Mark PJ, Ingram DM, Minchin RF
and Ratajczak T: Expression of the estrogen receptor-associated
immunophilins, cyclophilin 40 and FKBP52, in breast cancer. Breast
Cancer Res Treat. 58:267–280. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Desmetz C, Bascoul-Mollevi C, Rochaix P,
Lamy PJ, Kramar A, Rouanet P, Maudelonde T, Mangé A and Solassol J:
Identification of a new panel of serum autoantibodies associated
with the presence of in situ carcinoma of the breast in younger
women. Clin Cancer Res. 15:4733–4741. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Li G, Zhao F and Cui Y: Proteomics using
mammospheres as a model system to identify proteins deregulated in
breast cancer stem cells. Curr Mol Med. 13:459–463. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Connolly JL: Changes and problematic areas
in interpretation of the AJCC cancer staging manual, 6th edition,
for breast cancer. Arch Pathol Lab Med. 130:287–291.
2006.PubMed/NCBI
|
|
18
|
Singletary SE, Allred C, Ashley P, Bassett
LW, Berry D, Bland KI, Borgen PI, Clark G, Edge SB, Hayes DF, et
al: Revision of the American Joint Committee on Cancer staging
system for breast cancer. J Clin Oncol. 20:3628–3636. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Hwang SB, Bae JW, Lee HY and Kim HY:
Circulating tumor cells detected by RT-PCR for CK-20 before surgery
indicate worse prognostic impact in triple-negative and HER2
subtype breast cancer. J Breast Cancer. 15:34–42. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Miller DV, Leontovich AA, Lingle WL, Suman
VJ, Mertens ML, Lillie J, Ingalls KA, Perez EA, Ingle JN, Couch FJ
and Visscher DW: Utilizing nottingham prognostic index in
microarray gene expression profiling of breast carcinomas. Mod
Pathol. 17:756–764. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Elston CW and Ellis IO: Pathological
prognostic factors in breast cancer. I. The value of histological
grade in breast cancer: Experience from a large study with
long-term follow-up. Histopathology. 19:403–410. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Pereira H, Pinder SE, Sibbering DM, Galea
MH, Elston CW, Blamey RW, Robertson JF and Ellis IO: Pathological
prognostic factors in breast cancer. IV: Should you be a typer or a
grader ? A comparative study of two histological prognostic
features in operable breast carcinoma. Histopathology. 27:219–226.
1995. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Parham DM: Mitotic activity and
histological grading of breast cancer. Pahtol Annu. 30:189–207.
1995.
|
|
24
|
Patel RM and Folpe AL:
Immunohistochemistry for human telomerase reverse transcriptase
catalytic subunit (hTERT): A study of 143 benign and malignant soft
tissue and bone tumours. Pathology. 41:527–532. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Han YP, Ma CK, Wang SQ, Enomoto A, Zhao Y,
Takahashi M and Ma J: Evaluation of osteopontin as a potential
biomarker for central nervous system embryonal tumors. J
Neurooncol. 119:343–351. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Clarke C, Madden SF, Doolan P, Aherne ST,
Joyce H, O'Driscoll L, Gallagher WM, Hennessy BT, Moriarty M, Crown
J, et al: Correlating transcriptional networks to breast cancer
survival: A large-scale coexpression analysis. Carcinogenesis.
34:2300–2308. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ni Pau IB, Zakaria Z, Muhammad R, Abdullah
N, Ibrahim N, Emran Aina N, Abdullah Hisham N and Hussain Syed SN:
Gene expression patterns distinguish breast carcinomas from normal
breast tissues: The Malaysian context. Pathol Res Pract.
206:223–228. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kretschmer C, Sterner-Kock A, Siedentopf
F, Schoenegg W, Schlag PM and Kemmner W: Identification of early
molecular markers for breast cancer. Mol Cancer. 10:152011.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Győrffy B, Surowiak P, Budczies J and
Lánczky A: Online survival analysis software to assess the
prognostic value of biomarkers using transcriptomic data in
non-small-cell lung cancer. PLoS One. 8:e822412013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Li XP, Cao GW, Sun Q, Yang C, Yan B, Zhang
MY, Fu YF and Yang LM: Cancer incidence and patient survival rates
among the residents in the Pudong New Area of Shanghai between 2002
and 2006. Chin J Cancer. 32:512–519. 2013.PubMed/NCBI
|
|
31
|
Zeng H, Zheng R, Guo Y, Zhang S, Zou X,
Wang N, Zhang L, Tang J, Chen J, Wei K, et al: Cancer suevival in
China, 2003–2005: A population-based study. Int J Cancer.
136:1921–1930. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Gautier L, Cope L, Bolstad BM and Irizarry
RA: affy-analysis of Affymetrix GeneChip data at the probe level.
Bioinformatics. 20:307–315. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
De Leon JT, Iwai A, Feau C, Garcia Y,
Balsiger HA, Storer CL, Suro RM, Garza KM, Lee S, Kim YS, et al:
Targeting the regulation of androgen receptor signaling by the heat
shock protein 90 cochaperone FKBP52 in prostate cancer cells. Proc
Natl Acad Sci USA. 108:11878–11883. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Cheung-Flynn J, Prapapanich V, Cox MB,
Riggs DL, Suarez-Quian C and Smith DF: Physiological role for the
cochaperone FKBP52 in androgen receptor signaling. Mol Endocrinol.
19:1654–1666. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Hirota Y, Tranguch S, Daikoku T, Hasegawa
A, Osuga Y, Taketani Y and Dey SK: Deficiency of immunophilin
FKBP52 promotes endometriosis. Am J Pathol. 173:1747–1757. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Kumar P, Mark PJ, Ward BK, Minchin RF and
Ratajczak T: Estradiol-regulated expression of the immunophilins
cyclophilin 40 and FKBP52 in MCF-7 breast cancer cells. Biochem
Biophys Res Commun. 284:219–225. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Solassol J, Mange A and Maudelonde T: FKBP
family proteins as promising new biomarkers for cancer. Curr Opin
Pharmacol. 11:320–325. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Ott M, Litzenburger UM, Rauschenbach KJ,
Bunse L, Ochs K, Sahm F, Pusch S, Opitz CA, Blaes J, von Deimling
A, et al: Suppression of TDO-mediated tryptophan catabolism in
glioblastoma cells by a steroid-responsive FKBP52-dependent
pathway. Glia. 63:78–90. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Yang WS, Moon HG, Kim HS, Choi EJ, Yu MH,
Noh DY and Lee C: Proteomic approach reveals FKBP4 and S100A9 as
potential prediction markers of therapeutic response to neoadjuvant
chemotherapy in patients with breast cancer. J Proteome Res.
11:1078–1088. 2012. View Article : Google Scholar : PubMed/NCBI
|