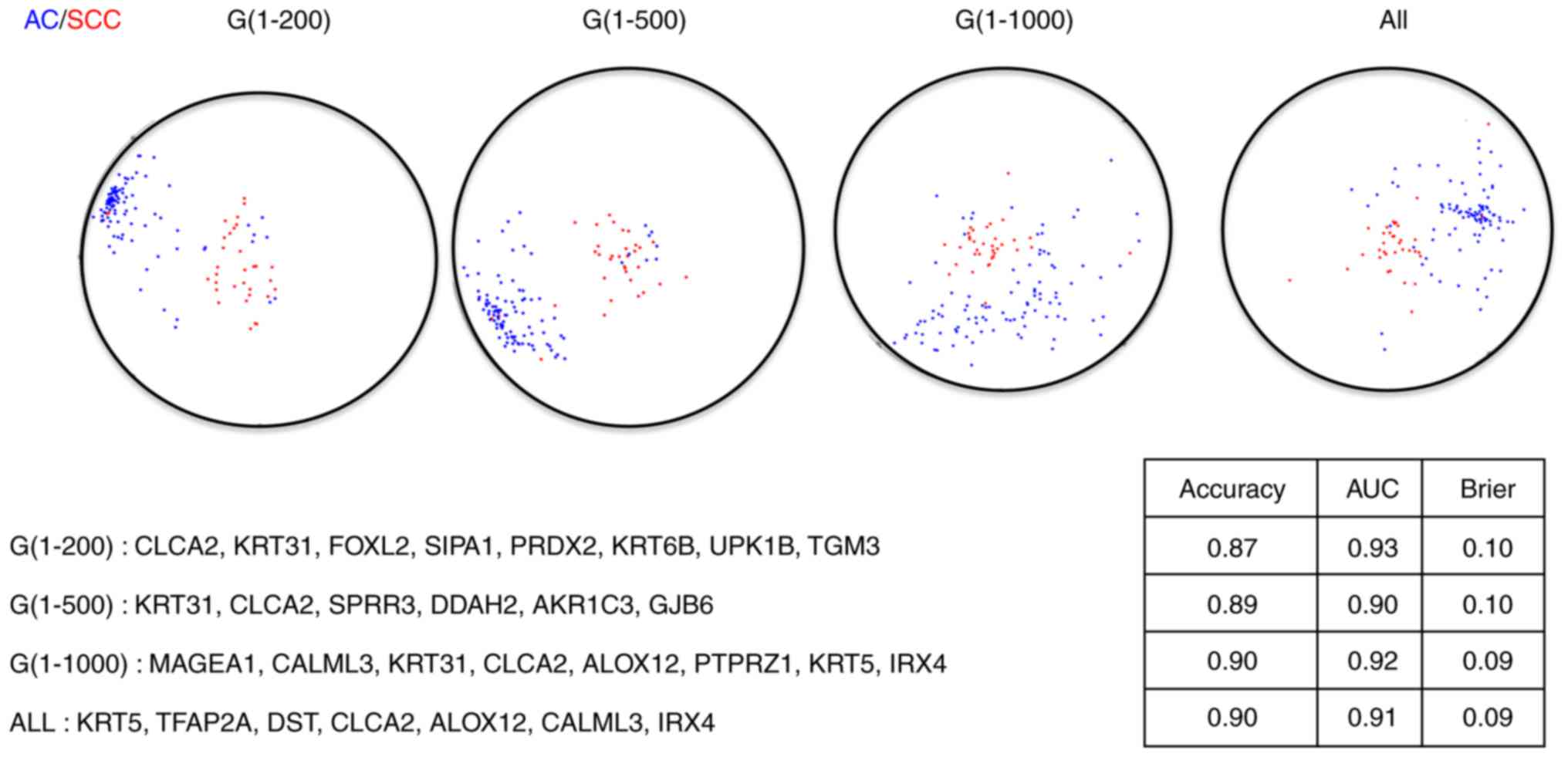
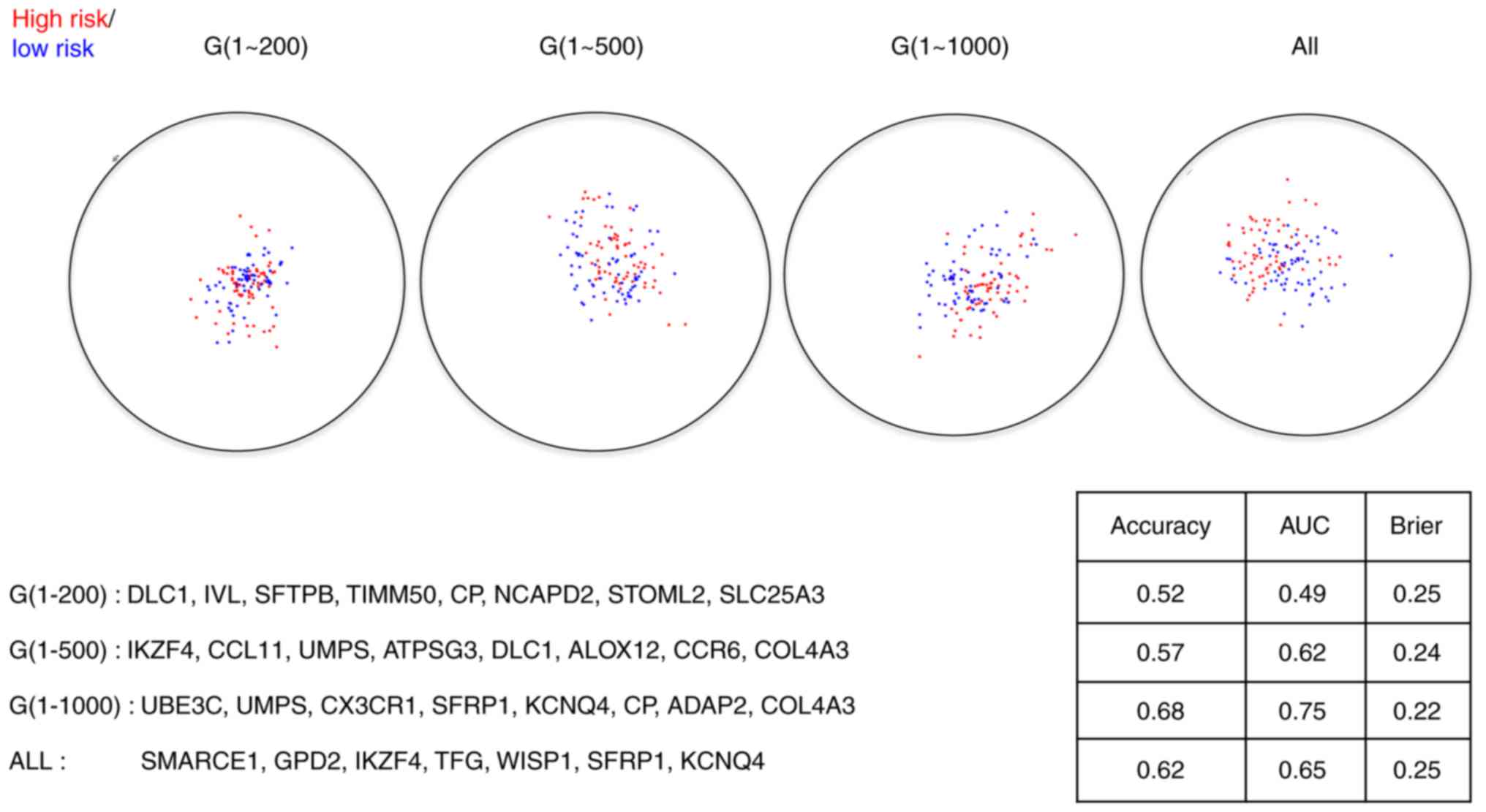
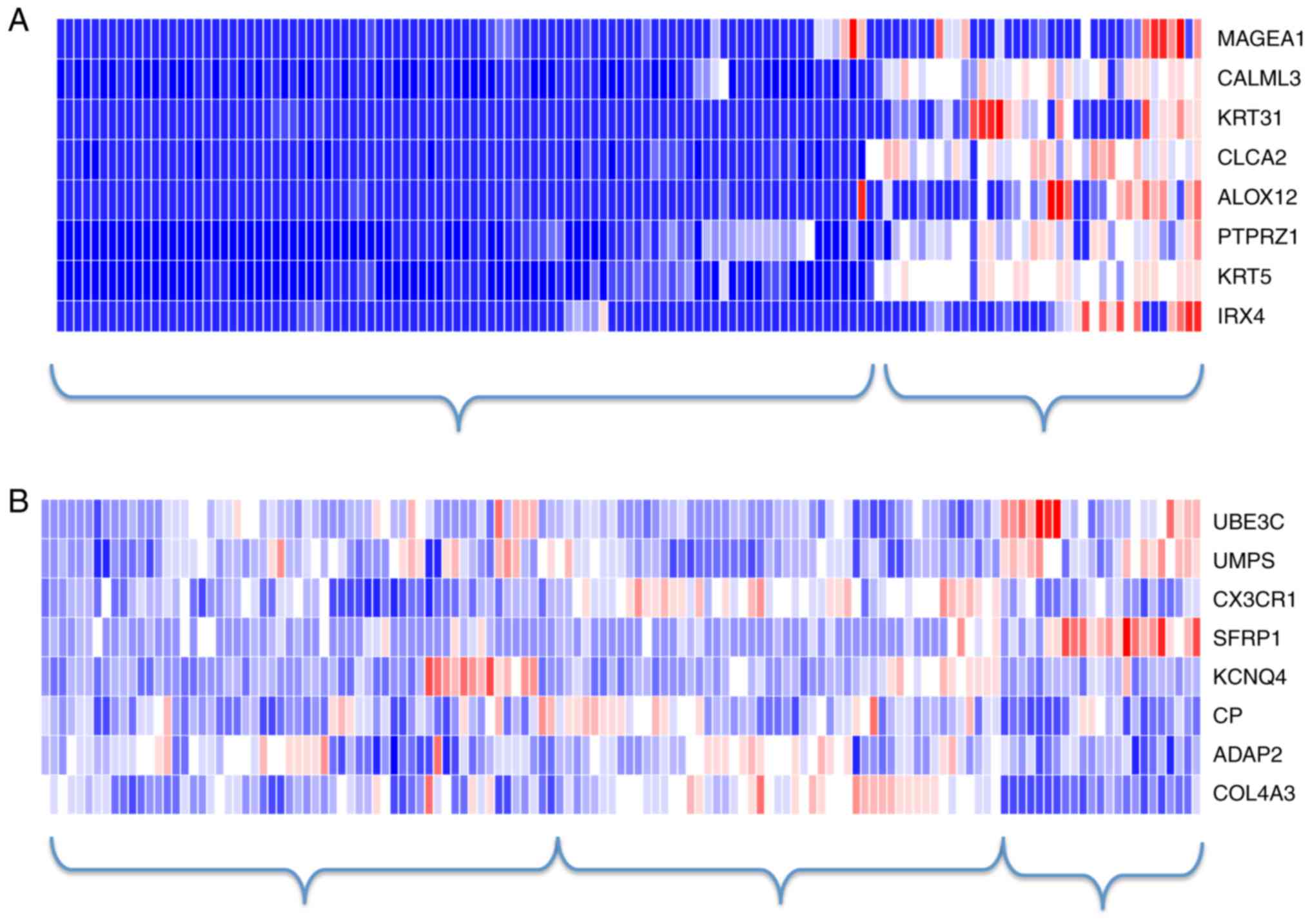
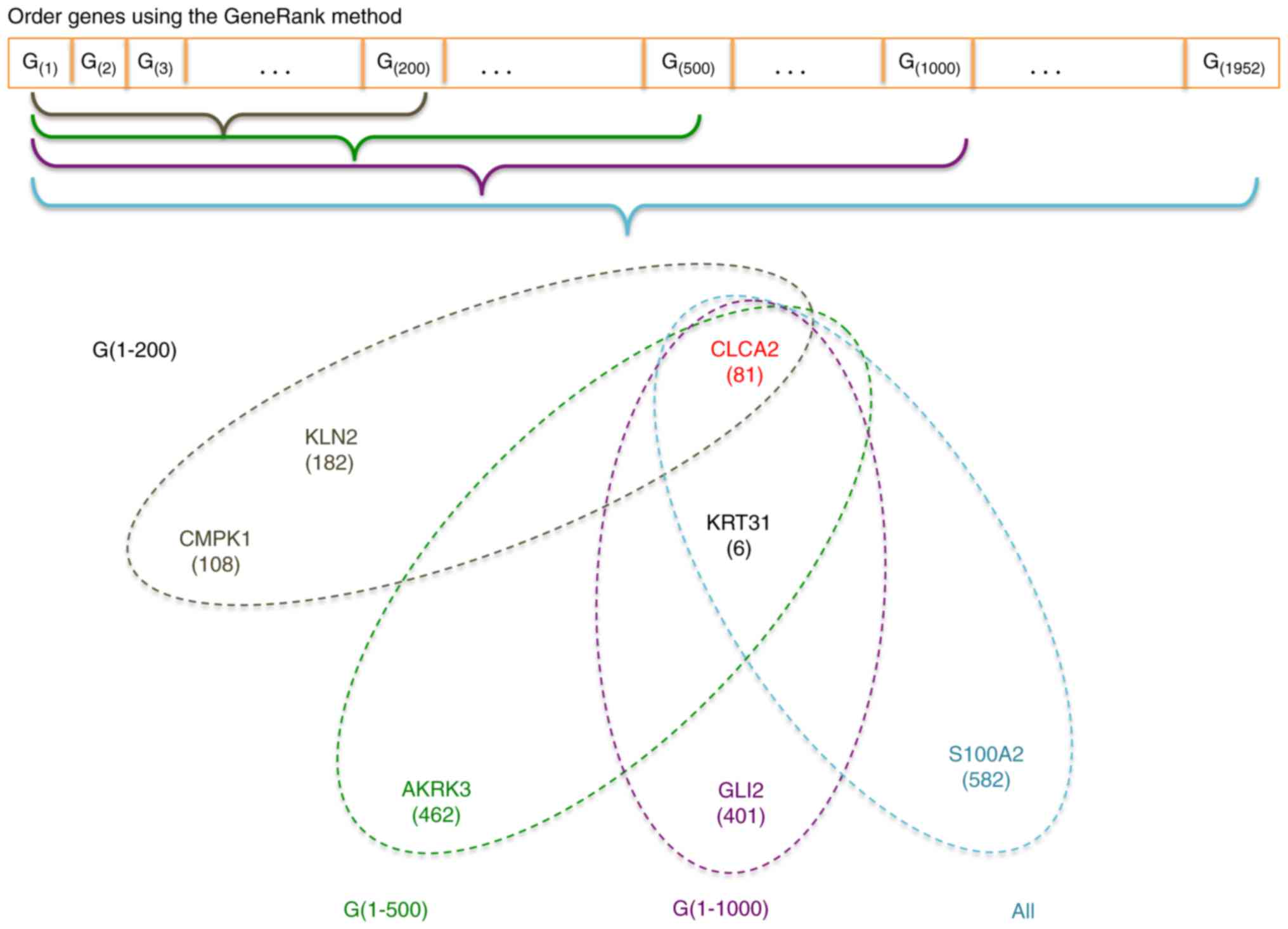
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Lu C, Onn A, Vaporciyan A, et al: 78:
Cancer of the lungHolland-Frei Cancer Medicine. 8th. People's
Medical Publishing House; 2010
|
|
3
|
Hou J, Aerts J, den Hamer B, van Ijcken W,
den Bakker M, Riegman P, van der Leest C, van der Spek P, Foekens
JA, Hoogsteden HC, et al: Gene expression-based classification of
non-small cell lung carcinomas and survival prediction. PLoS One.
5:e103122010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Tian S, Wang C and An MW: Test on
existence of histology subtype-specific prognostic signatures among
early stage lung adenocarcinoma and squamous cell carcinoma
patients using a Cox-model based filter. Biol Direct. 10:152015.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Tian S: Identification of subtype-specific
prognostic genes for early-stage lung adenocarcinoma and squamous
cell carcinoma patients using an embedded feature selection
algorithm. PLoS One. 10:e01346302015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Skrzypski M, Dziadziuszko R, Jassem E,
Szymanowska-Narloch A, Gulida G, Rzepko R, Biernat W, Taron M,
Jelitto-Górska M, Marjański T, et al: Main histologic types of
non-small-cell lung cancer differ in expression of
prognosis-related genes. Clin Lung Cancer. 14:666–673.e2. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Saeys Y, Inza I and Larrañaga P: A review
of feature selection techniques in bioinformatics. Bioinformatics.
23:2507–2517. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhang L, Wang L, Du B, Wang T, Tian P and
Tian S: Classification of non-small cell lung cancer using
significance analysis of microarray-gene set reduction algorithm.
Biomed Res Int. 2016:24916712016.PubMed/NCBI
|
|
9
|
Zhang A, Wang C, Wang S, Li L, Liu Z and
Tian S: Visualization-aided classification ensembles discriminate
lung adenocarcinoma and squamous cell carcinoma samples using their
gene expression profiles. PLoS One. 9:e110522014.
|
|
10
|
Tian S and Suárez-fariñas M:
Hierarchical-TGDR: Combining biological hierarchy with a
regularization method for multi-class classification of lung cancer
samples via high-throughput gene-expression data. Syst Biomed.
4:e259792013.
|
|
11
|
Ben-Hamo R, Boue S, Martin F, Talikka M
and Efroni S: Classification of lung adenocarcinoma and squamous
cell carcinoma samples based on their gene expression profile in
the sbv IMPROVER diagnostic signature challenge. Syst Biomed.
1:83–92. 2013.
|
|
12
|
Liu J, Yang XY and Shi WJ: Identifying
differentially expressed genes and pathways in two types of
non-small cell lung cancer: Adenocarcinoma and squamous cell
carcinoma. Genet Mol Res. 13:95–102. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Johannes M, Brase JC, Fröhlich H, Gade S,
Gehrmann M, Fälth M, Sültmann H and Beissbarth T: Integration of
pathway knowledge into a reweighted recursive feature elimination
approach for risk stratification of cancer patients.
Bioinformatics. 26:2136–2144. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Tian S, Chang HH and Wang C:
Weighted-SAMGSR: Combining significance analysis of microarray-gene
set reduction algorithm with pathway topology-based weights to
select relevant genes. Biol Direct. 11:502016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Chen L, Xuan J, Riggins RB, Clarke R and
Wang Y: Identifying cancer biomarkers by network-constrained
support vector machines. BMC Syst Biol. 5:1612011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Sun H, Lin W, Feng R and Li H:
Network-regularized high-dimensional Cox regression for analysis of
genomic data. Stat Sin. 24:1433–1459. 2014.PubMed/NCBI
|
|
17
|
Pan W, Xie B and Shen X: Incorporating
predictor network in penalized regression with application to
microarray data. Biometrics. 66:474–484. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Sokolov A, Carlin DE, Paull EO, Baertsch R
and Stuart JM: Pathway-based genomics prediction using generalized
elastic net. PLoS Comput Biol. 12:e10047902016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Hoffman P, Grinstein G, Marx K, Grosse I
and Stanley E: DNA visual and analytic data mining. Proceedings
Vis' 97. (Cat No 97CB36155). 1997.
|
|
20
|
Mramor M, Leban G, Demsar J and Zupan B:
Visualization-based cancer microarray data classification analysis.
Bioinformatics. 23:2147–2154. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Morrison JL, Breitling R, Higham DJ and
Gilbert DR: GeneRank: Using search engine technology for the
analysis of microarray experiments. BMC Bioinformatics. 6:2332005.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
McCall MN, Bolstad BM and Irizarry RA:
Frozen robust multiarray analysis (fRMA). Biostatistics.
11:242–253. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Law CW, Chen Y, Shi W and Smyth GK: Voom:
Precision weights unlock linear model analysis tools for RNA-seq
read counts. Genome Biol. 15:R292014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Leban G, Bratko I, Petrovic U, Curk T and
Zupan B: VizRank: Finding informative data projections in
functional genomics by machine learning. Bioinformatics.
21:413–414. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yeung KY, Bumgarner RE and Raftery AE:
Bayesian model averaging: Development of an improved multi-class,
gene selection and classification tool for microarray data.
Bioinformatics. 21:2394–2402. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Uno H, Cai T, Pencina MJ, D'Agostino RB
and Wei LJ: On the C-statistics for evaluating overall adequacy of
risk prediction procedures with censored survival data. Stat Med.
30:1105–1117. 2011.PubMed/NCBI
|
|
27
|
Zhao SD, Parmigiani G, Huttenhower C and
Waldron L: Más-o-menos: A simple sign averaging method for
discrimination in genomic data analysis. Bioinformatics.
30:3062–3069. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Binder H and Schumacher M: Comment on
‘network-constrained regularization and variable selection for
analysis of genomic data’. Bioinformatics. 24:2566–2569. 2008.
View Article : Google Scholar : PubMed/NCBI
|