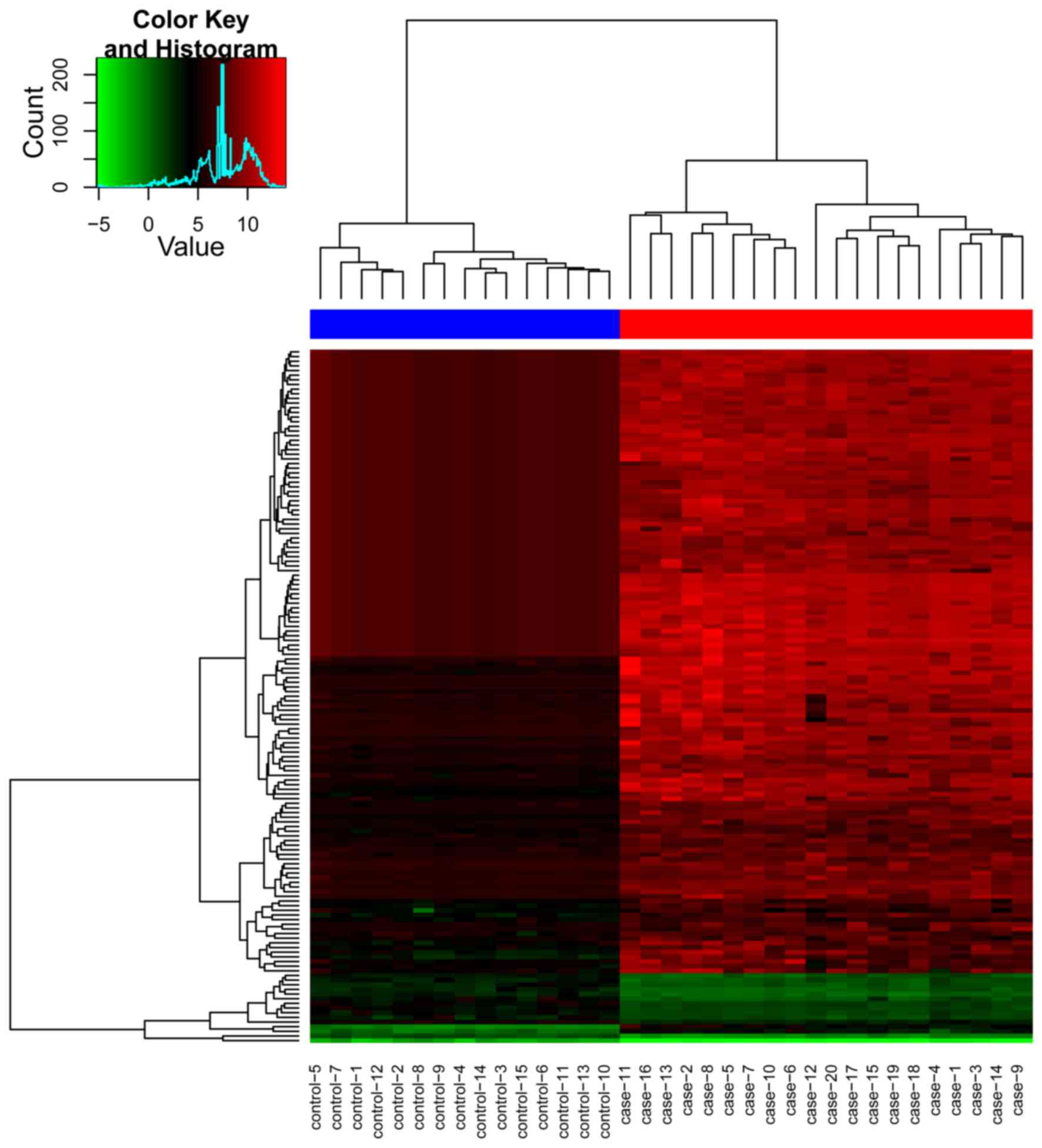
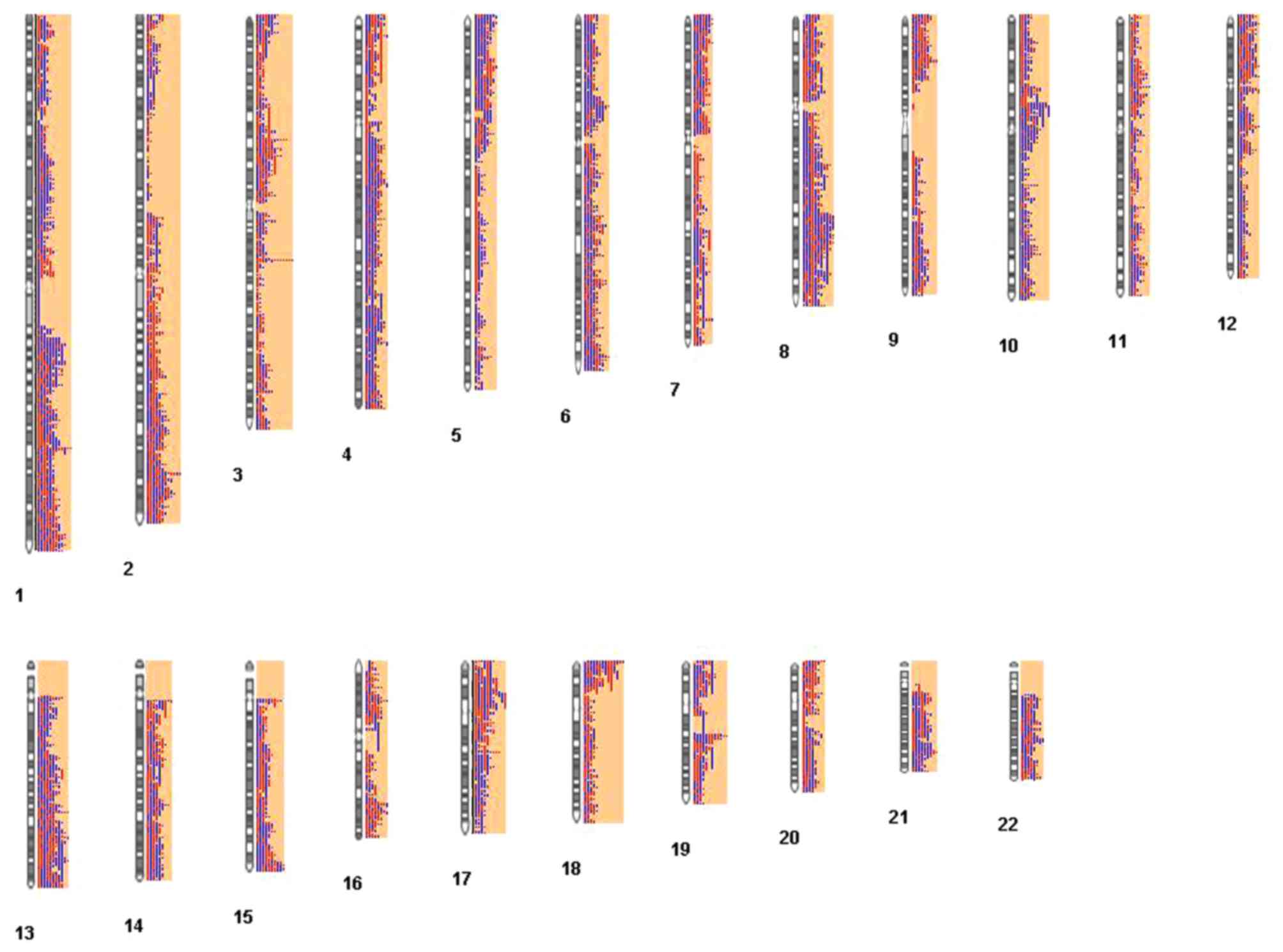
|
1
|
Savage SA, Mirabello L, Wang Z,
Gastier-Foster JM, Gorlick R, Khanna C, Flanagan AM, Tirabosco R,
Andrulis IL, Wunder JS, et al: Genome-wide association study
identifies two susceptibility loci for osteosarcoma. Nat Genet.
45:799–803. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kansara M and Thomas DM: Molecular
pathogenesis of osteosarcoma. DNA Cell Biol. 26:1–18. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Both J, Krijgsman O, Bras J, Schaap GR,
Baas F, Ylstra B and Hulsebos TJ: Focal chromosomal copy number
aberrations identify CMTM8 and GPR177 as new candidate driver genes
in osteosarcoma. PLoS One. 9:e1158352014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Broadhead ML, Clark JC, Myers DE, Dass CR
and Choong PF: The molecular pathogenesis of osteosarcoma: A
review. Sarcoma. 2011:9592482011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zong C, Lu S, Chapman AR and Xie XS:
Genome-wide detection of single-nucleotide and copy-number
variations of a single human cell. Science. 338:1622–1626. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Jin J, Cai L, Liu ZM and Zhou XS:
miRNA-218 inhibits osteosarcoma cell migration and invasion by
down-regulating of TIAM1, MMP2 and MMP9. Asian Pac J Cancer Prev.
14:3681–3684. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhang H, Wang Z, Bao L, et al: The
significance of copy number variation in multiple osteosarcoma's
malignance grade, drug resistance and classification. Mar Sci Bull.
24:41–46. 2015.
|
|
8
|
Cui W, Cai Y, Wang W, Liu Z, Wei P, Bi R,
Chen W, Sun M and Zhou X: Frequent copy number variations of
PI3K/AKT pathway and aberrant protein expressions of PI3K subunits
are associated with inferior survival in diffuse large B cell
lymphoma. J Transl Med. 12:102014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Silva AG, Krepischi AC, Pearson PL,
Hainaut P, Rosenberg C and Achatz MI: The profile and contribution
of rare germline copy number variants to cancer risk in Li-Fraumeni
patients negative for TP53 mutations. Orphanet J Rare Dis.
9:632014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Gokgoz N, Wunder JS and Andrulis IL:
Abstract 5075: Genome-wide analysis of DNA copy number variations
in osteosarcoma. Can Res. 72:50752012. View Article : Google Scholar
|
|
11
|
Conde L, Riby J, Zhang J, Bracci PM and
Skibola CF: Copy number variation analysis on a non-Hodgkin
lymphoma case-control study identifies an 11q25 duplication
associated with diffuse large B-cell lymphoma. PLoS One.
9:e1053822014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Shimada M: MicroRNA-mediated regulation of
apoptosis in osteosarcoma. J Carcinog Mutagen. 2013.doi:
10.4172/2157-2518.S6-001. View Article : Google Scholar
|
|
13
|
Jones KB, Salah Z, Del Mare S, Galasso M,
Gaudio E, Nuovo GJ, Lovat F, LeBlanc K, Palatini J, Randall RL, et
al: miRNA signatures associate with pathogenesis and progression of
osteosarcoma. Cancer Res. 72:1865–1877. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Osaki M, Takeshita F, Sugimoto Y, Kosaka
N, Yamamoto Y, Yoshioka Y, Kobayashi E, Yamada T, Kawai A, Inoue T,
et al: MicroRNA-143 regulates human osteosarcoma metastasis by
regulating matrix metalloprotease-13 expression. Mol Ther.
19:1123–1130. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Xu M, Jin H, Xu CX, Sun B, Song ZG, Bi WZ
and Wang Y: miR-382 inhibits osteosarcoma metastasis and relapse by
targeting Y box-binding protein 1. Mol Ther. 23:89–98. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wang Z and Cai H, Lin L, Tang M and Cai H:
Upregulated expression of microRNA-214 is linked to tumor
progression and adverse prognosis in pediatric osteosarcoma.
Pediatr Blood Cancer. 61:206–210. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Shimbo K, Miyaki S, Ishitobi H, Kato Y,
Kubo T, Shimose S and Ochi M: Exosome-formed synthetic microRNA-143
is transferred to osteosarcoma cells and inhibits their migration.
Biochem Biophys Res Commun. 445:381–387. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Salah Z, Arafeh R, Maximov V, Galasso M,
Khawaled S, Abou-Sharieha S, Volinia S, Jones KB, Croce CM and
Aqeilan RI: miR-27a and miR-27a* contribute to metastatic
properties of osteosarcoma cells. Oncotarget. 6:4920–4935. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhang H, Yin Z, Ning K, Wang L, Guo R and
Ji Z: Prognostic value of microRNA-223/epithelial cell transforming
sequence 2 signaling in patients with osteosarcoma. Hum Pathol.
45:1430–1436. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Huang YZ, Zhang J, Shao HY, Chen JP and
Zhao HY: MicroRNA-191 promotes osteosarcoma cells proliferation by
targeting checkpoint kinase 2. Tumour Biol. 36:6095–6101. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Fujiwara T, Katsuda T, Hagiwara K, Kosaka
N, Yoshioka Y, Takahashi RU, Takeshita F, Kubota D, Kondo T,
Ichikawa H, et al: Clinical relevance and therapeutic significance
of microRNA-133a expression profiles and functions in malignant
osteosarcoma-initiating cells. Stem Cells. 32:959–973. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Song QC, Shi ZB, Zhang YT, Ji L, Wang KZ,
Duan DP and Dang XQ: Downregulation of microRNA-26a is associated
with metastatic potential and the poor prognosis of osteosarcoma
patients. Oncol Rep. 31:1263–1270. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kobayashi E, Satow R, Ono M, Masuda M,
Honda K, Sakuma T, Kawai A, Morioka H, Toyama Y and Yamada T:
Microrna expression and functional profiles of osteosarcoma.
Oncology. 86:94–103. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Maire G, Martin JW, Yoshimoto M,
Chilton-MacNeill S, Zielenska M and Squire JA: Analysis of
miRNA-gene expression-genomic profiles reveals complex mechanisms
of microRNA deregulation in osteosarcoma. Cancer Genet.
204:138–146. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kuijjer ML, Rydbeck H, Kresse SH, Buddingh
EP, Lid AB, Roelofs H, Bürger H, Myklebost O, Hogendoorn PC,
Meza-Zepeda LA and Cleton-Jansen AM: Identification of osteosarcoma
driver genes by integrative analysis of copy number and gene
expression data. Genes Chromosomes Cancer. 51:696–706. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Camps C, Saini HK, Mole DR, Choudhry H,
Reczko M, Guerra-Assunção JA, Tian YM, Buffa FM, Harris AL,
Hatzigeorgiou AG, et al: Integrated analysis of microRNA and mRNA
expression and association with HIF binding reveals the complexity
of microRNA expression regulation under hypoxia. Mol Cancer.
13:282014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Lu X, Dai C, Hou A, Cui J, Cheng D and Xu
D: Dysregulated microRNA Profile in HeLa Cell Lines Induced by
LupeolBioinformatics Research and Applications. Springer; pp.
71–80. 2014
|
|
28
|
Frigerio CS, Lau P, Salta E, Tournoy J,
Bossers K, Vandenberghe R, Wallin A, Bjerke M, Zetterberg H,
Blennow K and De Strooper B: Reduced expression of hsa-miR-27a-3p
in CSF of patients with Alzheimer disease. Neurology. 81:2103–2106.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Chuang MK, Chiu YC, Chou WC, Hou HA,
Chuang EY and Tien HF: A simple, powerful, and widely applicable
Micro-RNA scoring system in prognostication of de novo myeloid
leukemia patients. Blood. 124:712014.
|
|
30
|
Chuang MK, Chiu YC, Chou WC, Hou HA,
Chuang EY and Tien HF: A 3-microRNA scoring system for
prognostication in de novo acute myeloid leukemia patients.
Leukemia. 29:1051–1059. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wang L, Zhu MJ, Ren AM, Wu HF, Han WM, Tan
RY and Tu RQ: A ten-microRNA signature identified from a
genome-wide microRNA expression profiling in human epithelial
ovarian cancer. PLoS One. 9:e964722014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Hirata H, Ueno K, Shahryari V, Deng G,
Tanaka Y, Tabatabai ZL, Hinoda Y and Dahiya R: MicroRNA-182-5p
promotes cell invasion and proliferation by down regulating FOXF2,
RECK and MTSS1 genes in human prostate cancer. PLoS One.
8:e555022013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Hirata H, Ueno K, Shahryari V, Tanaka Y,
Tabatabai ZL, Hinoda Y and Dahiya R: Oncogenic miRNA-182-5p targets
Smad4 and RECK in human bladder cancer. PLoS One. 7:e510562012.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zhang K, Chu K, Wu X, Gao H, Wang J, Yuan
YC, Loera S, Ho K, Wang Y, Chow W, et al: Amplification of FRS2 and
activation of FGFR/FRS2 signaling pathway in high-grade
liposarcoma. Cancer Res. 73:1298–1307. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Tania V, Ajay J, Naveen K, Steve D, Susan
M and Gnanapragasam VJ: Role and expression of FRS2 and FRS3 in
prostate cancer. BMC Cancer. 11:4842011. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Luo LY, Kim E, Cheung HW, Weir BA, Dunn
GP, Shen RR and Hahn WC: The tyrosine kinase adaptor protein FRS2
is oncogenic and amplified in high-grade serous ovarian cancer. Mol
Cancer Res. 13:502–509. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Valencia T, Joseph A, Kachroo N, Darby S,
Meakin S and Gnanapragasam VJ: Role and expression of FRS2 and FRS3
in prostate cancer. BMC Cancer. 11:4842011. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Wang J, Tsouko E, Jonsson P, Bergh J,
Hartman J, Aydogdu E and Williams C: Abstract P4-07-12: miR-206
inhibits cell migration through direct targeting of the
actin-binding protein coronin 1C in triple-negative breast cancer.
Cancer Res. 73 (24 Suppl):(P4): 07–12. 2013.
|
|
39
|
Mataki H, Enokida H, Chiyomaru T, Mizuno
K, Matsushita R, Goto Y, Nishikawa R, Higashimoto I, Samukawa T,
Nakagawa M, et al: Downregulation of the microRNA-1/133a cluster
enhances cancer cell migration and invasion in lung-squamous cell
carcinoma via regulation of Coronin1C. J Hum Genet. 60:53–61. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Novello C, Pazzaglia L, Cingolani C, Conti
A, Quattrini I, Manara MC, Tognon M, Picci P and Benassi MS: miRNA
expression profile in human osteosarcoma: Role of miR-1 and
miR-133b in proliferation and cell cycle control. Int J Oncol.
42:667–675. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Zhang C, Yao C, Li H, Wang G and He X:
Serum levels of microRNA-133b and microRNA-206 expression predict
prognosis in patients with osteosarcoma. Int J Clin Exp Pathol.
7:4194–4203. 2014.PubMed/NCBI
|
|
42
|
Bao YP, Yi Y, Peng LL, Fang J, Liu KB, Li
WZ and Luo HS: Roles of microRNA-206 in osteosarcoma pathogenesis
and progression. Asian Pac J Cancer Prev. 14:3751–3755. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
van Boxtel R, Gomez-Puerto C, Mokry M,
Eijkelenboom A, van der Vos KE, Nieuwenhuis EE, Burgering BM, Lam
EW and Coffer PJ: FOXP1 acts through a negative feedback loop to
suppress FOXO-induced apoptosis. Cell Death Differ. 20:1219–1229.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Santo EE, Ebus ME, Koster J, Schulte JH,
Lakeman A, van Sluis P, Vermeulen J, Gisselsson D, Øra I, Lindner
S, et al: Oncogenic activation of FOXR1 by 11q23 intrachromosomal
deletion-fusions in neuroblastoma. Oncogene. 31:1571–1581. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Xu H and Liu B: CPEB4 is a candidate
biomarker for defining metastatic cancers and directing
personalized therapies. Med Hypotheses. 81:875–877. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Ortiz-Zapater E, Pineda D, Martínez-Bosch
N, Fernández-Miranda G, Iglesias M, Alameda F, Moreno M, Eliscovich
C, Eyras E, Real FX, et al: Key contribution of CPEB4-mediated
translational control to cancer progression. Nat Med. 18:83–90.
2011. View Article : Google Scholar : PubMed/NCBI
|